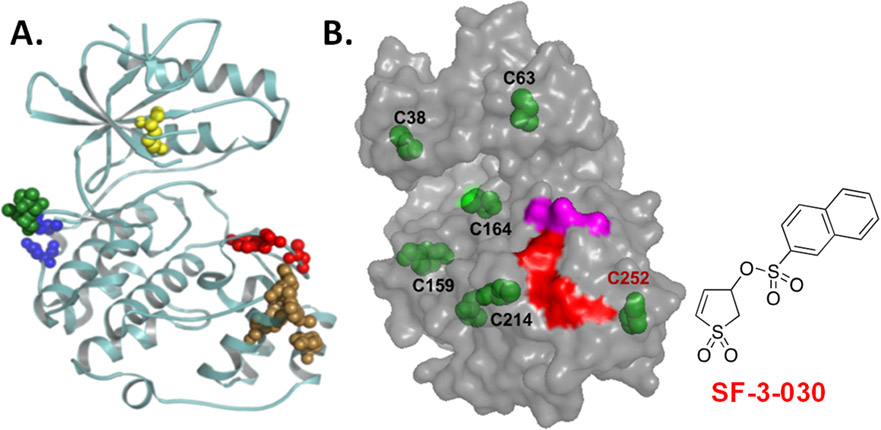
Figure 2.
(A) Ribbon structure of ERK2 (PDB:4GT3) highlights substrate docking sites. The D-domain recruitment site (DRS) or F-recruitment site (FRS) are indicated by blue/green or brown spheres, respectively. The conserved lysine found in the ATP binding site and the activation site are the yellow and red spheres, respectively. Unlike ATP binding/catalytic site inhibitors that block all kinase activity, function-selective ERK1/2 inhibitors target a select group of substrates that interact with the DRS or FRS. (B) SF-3-030 forms a cysteine adduct near the FRS. Model of ERK2 shows the position of all cysteine residues (green). High-resolution LC-MS identified C252 (red font) to be the primary (90%) residue modified by SF-3-030. The adjacent FRS (red) and activating TXY motif (magenta) are shown for reference.