Figure 7. Integrative modeling shows PEAR1 expression is a single, independent predictor of poor prognosis.
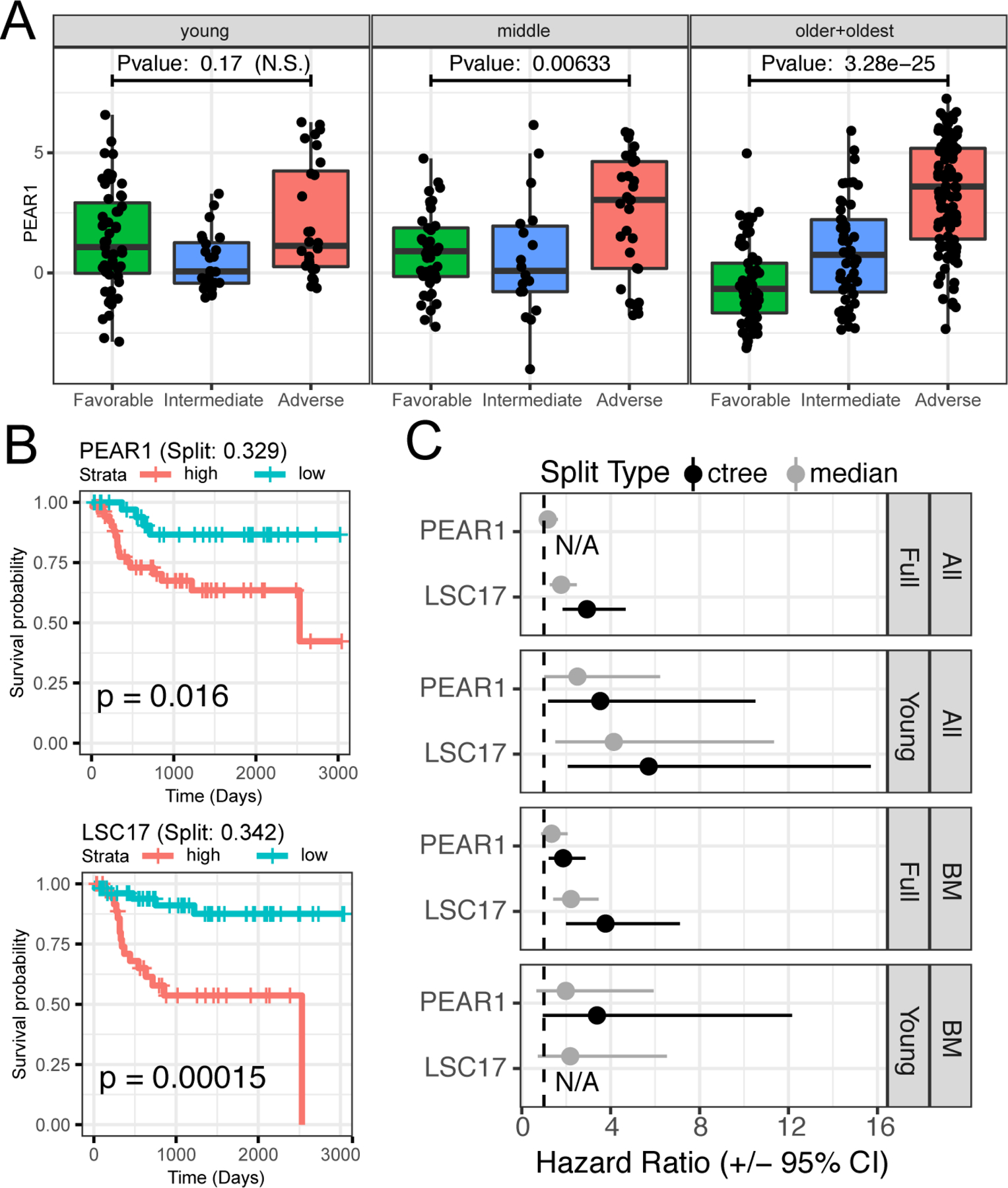
(A) PEAR1 expression (y-axis) is significantly higher in Adverse compared to Favorable ELN 2017 risk categorization (x-axis) for patients 60 and over (dots; older + oldest group), but the pattern is less pronounced in patients < 60. Significance determined using a using a Welch’s T-test. (B) PEAR1 expression differentiates survival for young patients equivalently to the LSC17 signature. Categorization of samples into high and low expression groups for PEAR1 and LSC17 was determined using the ctree methodology to facilitate comparison with PEAR1. (C) We compared performance of PEAR1 and LSC17 using both ctree and the median expression values for each. Shown are the hazard ratios (HRs) and 95% confidence intervals (CIs) for PEAR1 and LSC17 in the entire cohort or in the younger patients as well as in all sample types or only in bone marrow samples. The categories were high vs low expression as determined by either median threshold or significant splits using ctree (i.e., split type). In two instances, the ctree method failed to identify a split, and these are denoted with “N/A”.
