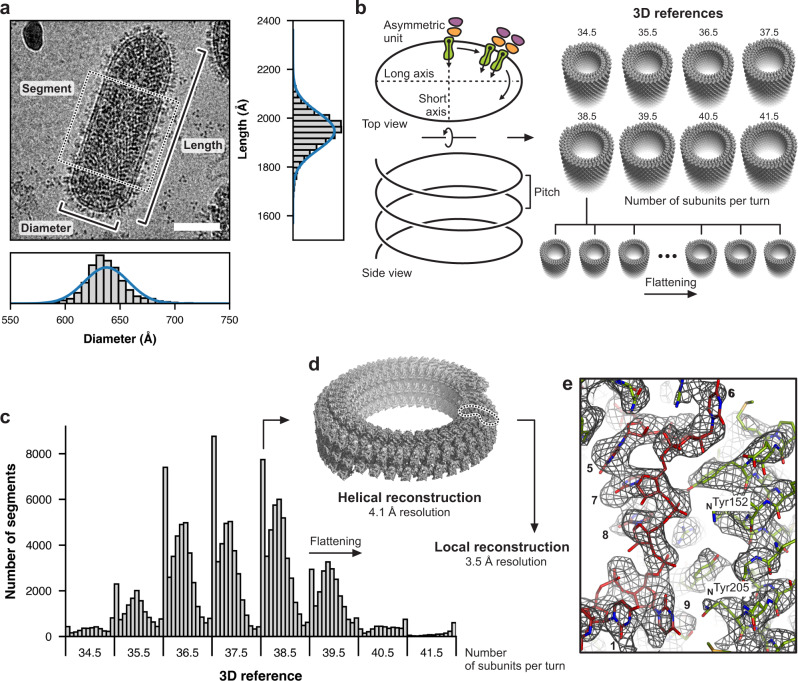
Fig. 1. Cryo-EM analysis of the helical VSV nucleocapsid.
a Cryo-EM image of a bullet-shaped vesicular stomatitis virus (VSV) particle. The image was low-pass filtered and contrast enhanced. The scale bar corresponds to 50 nm. The horizontal and vertical histograms show the distribution of measured diameter and length, respectively, from 5763 individual particles. Measurements were taken between the middle of the lipid bilayers as indicated and possible out-of-plane tilting was ignored. The solid blue lines are normal distribution fits to the histograms with means and standard deviations of 638 ± 20 Å for the diameter and 1947 ± 76 Å for the length. The dashed box shows one segment image extracted along the VSV trunk. b Preparation of 3D references for supervised classification. Density of one asymmetric unit initially obtained from a low resolution reconstruction comprising one N protein (green) and two M proteins (orange and purple) was expanded to create 3D references with different numbers of subunits per turn (34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, and 41.5) and different degrees of flattening. c Histogram of the supervised classification. Each segment image was assigned to one of the 96 3D references based on the highest alignment score. d Helical reconstruction obtained from sorted segments that scored best with un-flattened or only moderately flattened 3D references. Shown here is the reconstruction calculated from segments with 38.5 subunits per turn. e Close-up view of the cryo-EM density map after local reconstruction of the asymmetric unit with a nominal resolution of 3.5 Å. Subparticles were aligned by alignment-by-classification and subsequent local alignment. RNA bases are numbered.