Abstract
Introduction
Enamel hypoplasia causes a reduction in the thickness of affected enamel and is one of the most common dental anomalies. This defect is caused by environmental and/or genetic factors that interfere with tooth formation, emphasizing the importance of investigating enamel hypoplasia on an epidemiological and genetic level.
Methods
A genome-wide association of enamel hypoplasia was performed in multiple cohorts, overall comprising 7,159 individuals ranging in age from 7 to 82 years. Mixed models were used to test for genetic association while simultaneously accounting for relatedness and genetic population structure. Meta-analysis was then performed. More than 5 million single-nucleotide polymorphisms were tested in individual cohorts.
Results
Analyses of the individual cohorts and meta-analysis identified association signals close to genome-wide significance (p < 5 × 10<sup>–8</sup>), and many suggestive association signals (5 × 10<sup>–8</sup> < p < 5 × 10<sup>–6</sup>) near genes with plausible roles in tooth/enamel development.
Conclusion
The strongest association signal (p = 1.57 × 10<sup>–9</sup>) was observed near BMP2K in one of the individual cohorts. Additional suggestive signals were observed near genes with plausible roles in tooth development in the meta-analysis, such as SLC4A4 which can influence enamel hypoplasia. Additional human genetic studies are needed to replicate these results and functional studies in model systems are needed to validate our findings.
Keywords: Enamel hypoplasia, Dental anomalies, Genome-wide association studies, Multiethnicity, Genetics
Introduction
Tooth development (odontogenesis) is a complex process that starts early during embryogenesis and requires signaling between mesenchymal and epithelial tissues. Any disturbances during this complicated process in signaling pathways or mutations in any of the regulatory genes might cause dental anomalies, including changes in tooth morphology, structure, size, and number [1, 2]. Dental anomalies, such enamel hypoplasia, are assumed to be caused by interactions between environmental, genetic, and epigenetic factors during odontogenesis [3].
Tooth enamel is formed by ameloblasts, cells that produce and secrete enamel matrix proteins during the secretory stage of amelogenesis. There are two important groups of enamel proteins that are involved in the development of the enamel: the first group is non-amelogenins, which include enamelin (ENAM), ameloblastin (AMBN), tuftelin (TUFT1), and tuftelin-interacting protein 11 (TFIP11); the second group includes amelogenin (AMELX) [4].
Enamel hypoplasia is a quantitative defect that causes a reduction in the enamel thickness. This defect usually occurs during the secretory stage of amelogenesis and can affect both primary and permanent dentitions [5]. The clinical signs of enamel hypoplasia include shallow fossa (i.e., depression on the tooth surface) with either horizontal or vertical grooves and sometimes could lead to a partial or complete absence of enamel. Teeth affected with enamel hypoplasia could have an increased risk of dental caries [6, 7]. This is because enamel hypoplasia can create an enhanced environment for colonization, adhesion, and retention of cariogenic bacteria. Jointly with other dental caries risk factors, such as poor oral hygiene and cariogenic diet, dental caries might progress more rapidly [6]. Additionally, hypoplastic enamel has higher acid solubility than normal enamel, which makes it more susceptible to the acid secretions of cariogenic bacteria [8].
Several factors cause variation in the prevalence of enamel hypoplasia, such as: the method of examination, population studied, specific teeth, and dentition. The reported prevalence of enamel hypoplasia ranges between 0.8 and 17% in permanent dentition, while the prevalence of enamel hypoplasia in primary dentition ranges between 0.6 and 15.1% [9, 10, 11, 12]. Note that these variations in the prevalence of enamel hypoplasia may result from populations or dentition differences and/or the sample size available in each of these studies.
Genetic factors may contribute to the development of enamel hypoplasia, but to date there have been no large-scale gene-mapping attempts. Therefore, in an effort to better understand the genetic architecture of this relatively common dental trait, the aim of this study is to conduct genome-wide association studies (GWAS) to identify novel variants associated with enamel hypoplasia in four independent study cohorts and in all four cohorts combined via meta-analysis.
Materials and Methods
Study Sample
The participants for this study included individuals aged 7 years and older recruited through four studies: the Pittsburgh Orofacial Cleft Study (POFC, n = 3,579); the Center for Oral Health Research in Appalachia (COHRA1, n = 1,837; COHRA2, n = 1,195), a joint study of the University of Pittsburgh and West Virginia University; and the Pittsburgh Dental SCORE project (n = 548). The following is a basic description of each cohort.
Pittsburgh Orofacial Cleft Study
The first cohort for this study (n = 3,579) comes from the POFC. The POFC study populations were recruited from multiple cleft centers in the USA, including Colorado, Iowa, Pennsylvania, Texas, and Puerto Rico, and internationally, from Argentina, the Philippines, Colombia, Guatemala, and Hungary for the purpose of studying orofacial clefts. Recruitment of participants and the collection of the dental data for POFC started in 2006 and finished in 2016. The same data collection protocols were used for every site. This cohort included 1,392 control individuals, including control probands, their parents and siblings, and 2,187 unaffected relatives of individuals with orofacial clefts. Enamel hypoplasia data were collected via in-person dental exams, performed by trained and calibrated dentists or dental hygienists, and/or intraoral photographs scored by one of three calibrated dentists. The Modified Developmental Defects of Enamel Index [13] was used to diagnose and identify enamel hypoplasia and distinguish it from other enamel defects. Enamel hypoplasia can be defined as a quantitative defect in enamel, which was used for the identification of enamel hypoplasia. A visual dictionary was provided to every rater for the intraoral photographs (see online suppl. file 1; for all online suppl. material, see www.karger.com/doi/10.1159/000522642). The visual dictionary also included definition and photographs of other dental anomalies, such as hypocalcification, to help the rater distinguish enamel hypoplasia from the other dental anomalies. Enamel hypoplasia was recorded as present or absent for each tooth. Training and calibration for the intra-oral photos and the in-person dental exam was done at the University of Pittsburgh for all sites before the start of data collection. The three photo raters (B.J.H., L.M.M.U., and A.R.V.) each rated 15 randomly selected participants twice for calibration. Calibration was measured by comparing the enamel hypoplasia results from the ratings by L.M.M.U. and A.R.V. against the gold-standard rater, B.J.H. (B.J.H.'s intra-rater reliability [kappa] = 0.95). The Inter-rater reliability (kappa) across the 3 raters ranged between 0.91 and 0.93. Data from 15 participants who had both intra-oral photos and in-person dental exams were used to assess the reliability between those modalities, and the results showed good agreement between the two modes of assessment (kappa >80%) [14].
COHRA1
Our second cohort (n = 1,837) comes from the Center for Oral Health Research in Appalachia (COHRA), which was designed to address oral health disparities in the Appalachian region in the USA. Appalachia has the largest burden of oral health problems per capita in the USA. The participants in COHRA1 were recruited from rural Appalachia in West Virginia and Western Pennsylvania by using a household-based recruitment protocol and using the same data collection protocols for every site. The recruitment and data collection process, including the dental data, started in 2003 and ended in 2009. Training and calibration for the periodontal and dental exam was done at the University of Pittsburgh for all sites at the beginning of the data collection and periodically during the collection of the data. The dental examination was performed by trained and calibrated dental examiners (either a dentist or dental hygienist plus an assistant). The assessments were calibrated at the start of the study and during the course of data collection. The mean inter-and intra-rater reliability of the dental assessments, which included enamel hypoplasia, were excellent, i.e., 0.83 and 0.98, respectively [15].
COHRA2
The third cohort (COHRA2, n = 1,195) recruited women from West Virginia and southwest Pennsylvania who were in their first and second trimesters of pregnancy and followed them and their offspring until the children reached age 6 years. Data were collected through multiple in-person interviews, periodic telephone questionnaires, samples for DNA extraction, and dental assessment. The recruitment and data collection process, including the dental data, started in 2011 and is still ongoing. The dental examination was performed by trained and calibrated dental examiners (either a dentist or dental hygienist). The same definition and index of enamel hypoplasia in POFC, Modified Developmental Defects of Enamel Index [13], was used here and enamel hypoplasia was recorded as absent or present for each tooth. The calibration and training on caries assessment was done before the beginning of the data collection and on a regular basis. The intra-rater reliability of dental caries assessments ranged between 46.1 and 80.6. The inter-rater reliability of dental caries assessments showed a good agreement with kappa = 87.8 [16].
Dental SCORE
The fourth cohort (Dental SCORE, n = 548) is a subset of a larger project, Heart SCORE, which is an ongoing cardiovascular disease study of 2,000 middle aged White and African Americans in the Pittsburgh region of Pennsylvania [17]. The recruitment and data collection process, including that of the dental data, started in 2007 and ended in 2010. In Dental SCORE, an extended dental and periodontal examination was performed following the same protocols as in the COHRA1 study. The dental examination was performed by trained and calibrated dental examiners (either a dentist or dental hygienist plus an assistant).
Table 1 provides a basic description of each cohort in regard to age, sex, enamel hypoplasia, and the mean score of dental caries assessed by counts of the decayed and filled teeth due to caries (DFT) index. More details regarding data collection are provided in online supplementary Table S1.
Table 1.
Demographics of each cohort included in the study
| Male | Female | Total | Age, years | Enamel hypoplasia |
Most affected teeth with enamel hypoplasia | DFTa | ||
|---|---|---|---|---|---|---|---|---|
| no | yes | |||||||
| POFC | 1,505 (42.1) | 2,074 (57.9) | 3,579 (50) | 31±14.7 | 3,297 (92.1) | 282 (7.9) | Maxillary left central incisors (15.24%), maxillary right central incisors (14.89%), and maxillary right lateral incisors (14.89%) | 4.36±4.28 |
| COHRA1 | 783 (42.6) | 1,054 (57.4) | 1,837 (25.7) | 25.5±13.7 | 1,789 (97.4) | 48 (2.6) | Maxillary right central incisors (7.84%), maxillary right first molars (6.86), maxillary left lateral incisors (5.88%) | 5.96±5.11 |
| COHRA2 | 32 (2.7) | 1163 (97.3) | 1,195 (16.7) | 30.9±7.9 | 1,115 (93.3) | 80 (6.7) | Maxillary right first molar (6.60%), maxillary left first molar (6.13%), and maxillary left lateral incisors (5.18%) | 5.13±6.93 |
| Dental SCORE | 179 (32.7) | 369 (67.3) | 548 (7.6) | 63.1±7.5 | 451 (82.3) | 97 (17.7) | Maxillary right first molar (11.34%), maxillary right second molar (10.31%), and maxillary left lateral incisors (6.18%) | 12.07±5.33 |
| Total | 2,499 (34.9) | 4,660 (65.1) | 7,159 | 37.6±10.95 | 6,652 (92.9) | 507 (7.1) | 6.88±5.41 | |
Data are presented as n (%) or the mean ± SD.
DFT counts of decayed and filled teeth due to caries.
Enamel Hypoplasia Phenotypes
Trained and calibrated dental examiners (dentists or dental hygienists) performed the dental examination for the participants. Enamel hypoplasia in each study was scored for each tooth in the entire dentition except for the wisdom teeth. We looked at the distribution of which teeth were affected with enamel hypoplasia and found that the maxillary incisors (central and lateral) are the most affected teeth with enamel hypoplasia (see Table 1 for complete details). For the purpose of this study, enamel hypoplasia was analyzed as a binary trait, with values “0” when no teeth are affected with enamel hypoplasia, and “1” where one tooth or more is affected with enamel hypoplasia. The calibration and training information were discussed in the previous section.
DNA Collection, Genotyping, and Quality Control
All the sites in POFC and the COHRA2 studies used Oragene kits for DNA saliva sample collection. For COHRA1 and Dental SCORE, blood samples were the priority, however saliva (Oragene), cheek swab (Oragene), or mouthwash buccal cell samples were taken if blood samples were not possible to collect.
Genotyping for all cohorts was performed by the Johns Hopkins University Center for Inherited Disease Research (CIDR, https://cidr.jhmi.edu/). The genotyping chip used in COHRA1 was the Illumina Human610-Quadv1_B BeadChip (Illumina, San Diego, CA, USA) [18], while COHRA2, Dental SCORE, and a subset of COHRA1 were genotyped on the Illumina Multi-Ethnic Genotyping Array (MEGA) chip. For the POFC study the Illumina HumanCoreExome array with added custom content was used for genotyping [19].
The genetic data were imputed using IMPUTE2 [20] to a comprehensive imputation panel. The reference panel for POFC was the phase 3 reference panel from the 1000 Genomes Project. For COHRA the 1000 Genomes project phase 1 version 2 release was used [21]. Genotype data went through an extensive process of cleaning, imputation, and quality assurance, performed by CIDR and/or the CIDR Genetics Coordinating Center at the University of Washington. Standard quality-control criteria were applied to filter the SNPs. SNPs were excluded if they had (i) a missing call rate of ≥5% in cases or controls; (ii) >1 discordant calls; (iii) SNPs with MAF <1% in the population; (iv) significant deviation from Hardy–Weinberg equilibrium (p values less than 10−4); (v) >25 Mendelian errors; (vi) imputed genotypes were filtered out if their probability was <0.9; [22] SNPs with INFO score < 0.5; and (viii) more than 1 HapMap replicate error [19, 23, 24]. The total of SNPs that were available to analyze after applying the standard quality control measures were 6,025,027 for COHRA1, 5,375,488 for COHRA2, 5,375,490 for Dental SCORE, and 7,168,909 for POFC. Table S2 in the supplemental materials provides a summary of the DNA collection, storage, genotyping and dbGaP accession numbers for each project.
Statistical Methods
All descriptive and statistical analyses were performed using the R statistical analysis environment version 3.4.1 (R Foundation for Statistical Computing, Vienna, Austria; https://www.Rproject.org/). Associations between SNPs and the binary enamel hypoplasia trait were assessed using mixed models with adjustment for age, age2, sex, and recruitment site, carried out using Efficient Mixed-Model Association Expedited (EMMAX) software [25]. EMMAX was selected to account for relatedness and population structure [26].
A GWAS of enamel hypoplasia was conducted in each of the four independent cohorts and followed up with a meta-analysis. SNPs strongly associated with enamel hypoplasia were further evaluated for their likely function based on publicly available genomic annotation databases. In the meta-analysis, the results obtained from the four individual GWAS were combined, using Stouffer's p value-based meta-analysis method in the software package METAL [27]. The METAL heterogeneity test (I2) was used to find which observed effect sizes are heterogeneous across the four cohorts.
To account for multiple testing in GWAS results, a genome-wide significance threshold of p < 5 × 10−8 was applied, assuming 1 million independent tests. The p value threshold for suggestive significance was set to 5 × 10−8 < p < 5 × 10−6. R was also used to calculate the genomic inflation factor, lambda (λ), and to create Manhattan plots to visualize the association results. More details regarding the statistical methods are available in the supplementary materials.
Functional Annotation
Identification of genes of interest were based on physical proximity using ±500 kb windows from the lead SNP at each of the loci associated with enamel hypoplasia. These genes were queried for corroborating biological connections to tooth development, oral health, and/or craniofacial development in different online databases, including the National Center for Biotechnology Information (NCBI) databases, specifically the Gene, PubMed, and OMIM. In addition, HaploReg v.4.1 was used to acquire information on chromatin interaction, expression quantitative trait loci (eQTL), gene regulation, and linkage disequilibrium (LD) of the lead SNPs [28].
GWAS summary statistics from the meta-analysis were entered into the Functional Mapping and Annotation (FUMA) of Genome-Wide Association Studies (FUMA) platform, a downstream GWAS analysis tool, using default parameters to prioritize, annotate, and interpret the genomic variants and genes from the meta-analysis results [29].
Results
In the four cohorts combined, the majority of the participants were white (63.8 vs. 35.2% other races), there were more female participants than male participants (65.1 vs. 34.9%), and the mean age was 37.6 years. The overall prevalence of enamel hypoplasia significantly differed across the cohorts (p < 0.00001), with 7.9% of participants in POFC (n = 3,579), compared with 2.6% in COHRA1 (n = 1,837), 6.7% in COHRA2 (n = 1,195), and 17.7 % in Dental SCORE (n = 548). The prevalence of enamel hypoplasia among white participants across cohorts ranged from 2.3% in COHRA1 to 10.6% in Dental SCORE (p < 0.00001). According to previous studies that investigated enamel hypoplasia in comparable populations to ours (Brazil, Spain, and Australia), prevalence of enamel hypoplasia ranges from 0.6% to 11% in the primary dentition and from 0.8% to 17% in the permanent dentition. Thus, the prevalence of enamel hypoplasia in the current study is within the previously reported ranges [9, 10, 12].
Genome-Wide Association Studies
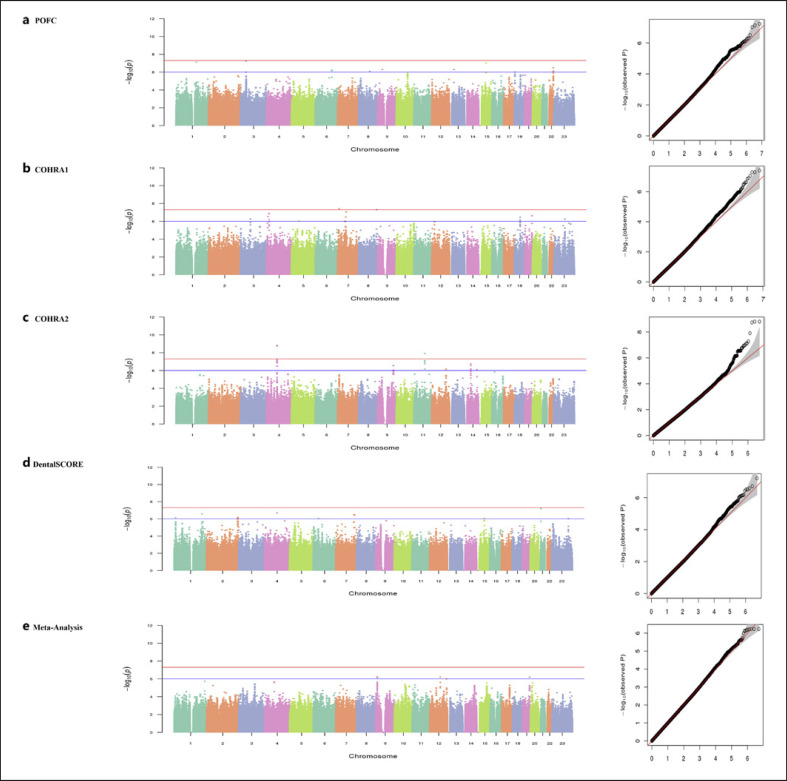
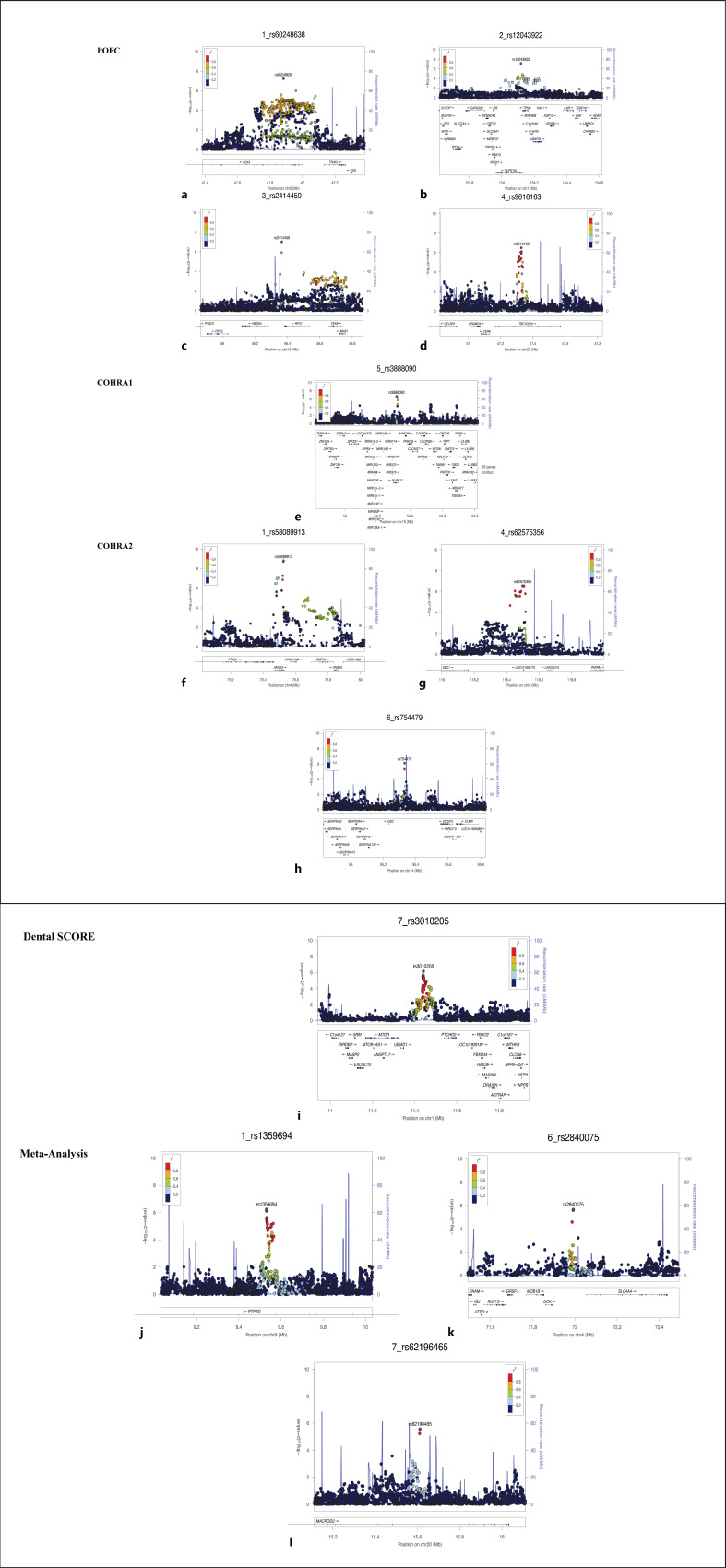
This section summarizes the GWAS results in each cohort and across all the cohorts combined by meta-analysis. A number of association signals were observed in the individual cohorts, both at the genome-wide and suggestive significance levels. Highlights from these results are presented below. No evidence of genomic inflation was observed in any of our independent GWASs or the meta-analysis (lambdas range between 0.99 and 1.02). Manhattan and quantile-quantile (Q-Q) plots of GWAS results for enamel hypoplasia for each cohort and meta-analysis are shown in Figure 1. Association results for the lead SNPs at associated loci with enamel hypoplasia are shown in Table 2. Furthermore, lead SNPs that showed association with enamel hypoplasia were further visualized using LocusZoom [30] plots in order to assess whether genes in such loci may have a possible role in tooth development and dental/oral health (Fig. 2).
Fig. 1.
a–e Manhattan plots and quantile-quantile (Q-Q) plots show GWAS results for the analyses. Red lines represent thresholds for genome-wide significance (p value <5 × 10−8). Blue lines represent thresholds for suggestive significance (p value <5 × 10−6).
Table 2.
GWAS results for enamel hypoplasia
| rsID | Chr | BP | Beta | p | A1/A2a | Nearby gene(s)b |
|---|---|---|---|---|---|---|
| POFC | ||||||
| rs60248638c | 3 | 41883692 | –0.0600436 | 5.87E–08 | A/G | ULK4, CCK |
| rs12043922c | 1 | 154118497 | –0.0741821 | 7.45E–08 | A/G | NUP210L, ADAR |
| rs2414459 | 15 | 56365357 | –0.0610045 | 9.58E–08 | C/T | PYGO1 |
| rs9616163c | 22 | 47328584 | –0.0683888 | 3.22E–07 | A/G | TBC1D22A, CELSR1 |
|
| ||||||
| COHRA1 | ||||||
| rs245664 | 7 | 9089477 | –0.0643447 | 3.92E–08 | C/G | NXPH1 |
| rs62522905 | 8 | 136395352 | –0.0504171 | 5.04E–08 | A/G | KHDRBS3 |
| rs6954583 | 7 | 63292425 | 0.05167179 | 8.91E–08 | C/T | LINC01005 |
| rs3888090c | 19 | 54317914 | –0.0647593 | 2.37E–07 | A/G | NLRP12 |
|
| ||||||
| COHRA2 | ||||||
| rs58089913c | 4 | 79526046 | –0.1514529 | 1.57E–09 | A/G | ANXA3, BMP2K |
| rs11232439 | 11 | 80743780 | –0.1037051 | 1.24E–08 | G/T | LOC101928944 |
| rs57555457 | 14 | 48260934 | –0.1046353 | 1.80E–07 | C/T | MIR548Y |
| rs62575356 | 9 | 118497768 | –0.1189798 | 2.77E–07 | C/T | LOC101928775, PAPPA |
| rs754479 | 14 | 95331008 | –0.1108255 | 8.01E–07 | C/T | GSC |
|
| ||||||
| Dental SCORE | ||||||
| rs2142491 | 21 | 16916381 | –0.2386754 | 6.18E–08 | C/T | USP25 |
| rs28504363d | 4 | 94836997 | –0.2762404 | 1.94E–07 | A/G | ATOH1 |
| rs36010081c | 1 | 212423782 | –0.2066991 | 2.67E–07 | A/C | PPP2R5A |
| rs7780210 | 7 | 139231244 | –0.1753172 | 3.06E–07 | C/G | CLEC2L |
| rs3010205 | 1 | 11440957 | 0.1198029 | 7.22E–07 | A/G | UBIAD1, MTOR |
A1 is the effect allele, A2 is the other allele.
Gene within the +500-kb window.
These SNPs were intronic.
This SNP is the only one that was genotyped, the rest of the SNPs in the table were imputed.
Fig. 2.
a–l LocusZoom of regions of interest. The genome build used for the recombination rate was based on 1,000 Genomes November 2014 EUR data. All of the gene positions and directions of transcription are annotated on the plots.
POFC GWAS
Several signals were near genes with potentially relevant biological roles to tooth development and enamel hypoplasia. One of the top hits involved SNP rs60248638 (p = 5.87 × 10−8; Fig. 2a) which is intronic to ULK4 (Unc-51-like kinase 4). ULK4 has been shown to have an essential role in regulating the wnt signaling pathway. This is specifically relevant as the wnt signaling pathway is a fundamental regulator of tooth development [31]. In addition, the wnt signaling pathway is crucial during brain development. Indeed, Ulk4 mutant mice appear to have severe reduction of cortex and neural stem cells, which indicates that embryonic neurogenesis is affected [32, 33]. Furthermore, a study that investigated, among other things, a rare deletion of 143,487 bp at Chr3:41611009-41754496 which encodes exons 25–30 of ULK4 in 6 patients found that these patients manifested heterogeneous clinical symptoms, including behavioral problems, developmental delay, learning difficulties, language delay, speech delay, and enamel hypoplasia [32]. These manifestations support the previous evidence that ULK4 has a role in brain development and/or function and could possibly hint at a possible role of ULK4 during odontogenesis (tooth development). Rs60248638 is also located approximately 400 kb downstream of CCK, which encodes a member of the gastrin/cholecystokinin family of proteins and has a role as a hormone and neuropeptide for multiple functions including the release of digestive enzymes from the pancreas and as an autocrine growth factor [34]. A previous genome-wide screening study evaluated enamel organ cells from the secretory and maturation stage demonstrated that CCK was highly up-regulated during enamel maturation [35]. Based on this supporting biological evidence, ULK4 and CCK may influence tooth development and/or enamel hypoplasia. However, their exact roles need to be investigated more to confirm their association with enamel hypoplasia.
The SNP rs12043922 (p = 7.45 × 10−8; Fig. 2b) is intronic to NUP210L and the function of this gene is still unknown. This SNP is also located approximately 400 kb downstream of ADAR, which produces RNA-specific adenosine deaminase 1 (ADAR1) protein. The expression of Adar1 is very strong in mice dental papilla, ameloblasts, and odontoblasts during the process of tooth development [36]. In a previous human study, a novel mutation in ADAR1 led to dental anomalies, specifically dens evaginatus and dens invaginatus [36]. The precise function of ADAR1 in tooth development is not yet clear; however, the previous study suggested that it might influence enamel hypoplasia and tooth development.
A suggestive association signal was detected near rs2414459 (p = 9.58 × 10−8; Fig. 2c), which is located approximately 400 kb upstream of PYGO1. Mice lacking both Pygo1 and Pygo2 in epithelial cells were able to develop teeth, yet the enamel had been structurally disorganized, were bright white, and had reduced iron content compared to control mice. These characteristics are very similar to the ones humans develop with amelogenesis imperfecta [37]. This SNP is in high LD with rs77220396 (r2 = 0.86), and the C allele of rs77220396 could change the affinity of Sox_2 (score 13–13.2). Sox2 has been found to be expressed during tooth development in mice at different stages [38, 39]. While it is still unclear how PYGO1 and SOX2 may influence the risk of developing enamel hypoplasia, there is biological evidence that both genes may impact tooth development and/or enamel hypoplasia.
The second top association signal with enamel hypoplasia was detected near rs9616163 (p = 3.22 × 10−7; Fig. 2d), which is intronic to TBC1D22A; however, the precise biological role of this gene is unknown. This SNP is in high LD with several SNPs that show enhancer chromatin marks in many different cell types, including osteoblasts (i.e., bone-forming cells), which can be found in periodontal ligament and alveolar bone in the oral cavity. rs9616163 is located 400 kp upstream of CELSR1. The protein encoded by CELSR1 belongs to the flamingo subfamily, which has a role during early embryogenesis. Celsr1 expression was analyzed alongside other genes, including Fzd3, Fzd6, Vangl2, Dvl2, and Dvl3, using mouse molar teeth from the bed to bell stage. Overall, Celsr1 was expressed in different tooth cells, including odontoblast and ameloblast, and during different tooth development stages. In addition, Celsr1 was found to be highly expressed in the enamel knot during the cap stage [40]. Further investigations are needed to clarify how CELSR1 may influence tooth development and/or enamel hypoplasia.
COHRA1 GWAS
The genome-wide significant loci in COHRA1 did not include genes with clear biological evidence for tooth development, enamel, oral health, and/or craniofacial development. For example, the top significant association signal was detected near rs245664 (p = 3.92 × 10−8) and this SNP is located 500 kb downstream to neurexophilin (NXPH1), which promotes adhesion between dendrites and axons and has been linked to neuroticism [41]. Both of the following most strongly associated variants (rs62522905 and rs6954583) are within 200 kb from genes (KHDRBS3, LINC01005), and both of these genes' biological functions are still unknown.
A suggestive variant identified was rs3888090 (p = 2.37 × 10−7; Fig. 2e), intronic to NLRP12, which is an atypical intracellular sensor of the NLR family and plays a role in negatively regulating several inflammatory conditions and osteoclastogenesis. A recent study that investigated the role of NLRP2 in the immune response and bone loss initiated by a bacterial infection in the oral cavity, more specifically during apical periodontitis, found that NLRP12 could help in reducing alveolar bone loss and dampening inflammatory response and osteoclastogenesis through the negative regulation of the NF-κB pathway [42]. Interestingly, this SNP shows enhancer chromatin marks in osteoblasts, which can be found in periodontal ligament and alveolar bone in the oral cavity, and that could indicate that variation in NLRP12 can be associated with periodontitis. It is unclear, though, how variation within this genomic locus could influence enamel hypoplasia and/or tooth development.
COHRA2 GWAS
The strongest association signal among the four cohorts was detected in this cohort at rs58089913 (p = 1.57 × 10−9; Fig. 2f). This SNP is intronic to ANXA3 (Annexin A3), which has a regulatory role in several biological process, such as containing cell proliferation, angiogenesis, and cancer progression [43]. This SNP is also located 200 kb upstream of BMP2K. The encoded protein of BMP2K plays a presumed role in the regulation of the mitigation of osteoblast differentiation, which makes BMP2K associated with the development of the bone and the cartilage [44, 45]. In fact, BMP2K is known as a BMP2-inducible gene [46]. BMP2 has a well-established role in early embryogenesis, skeletal development, and tooth development [47, 48, 49, 50]. Motif analysis showed that the A allele of rs58089913 could change the affinity of FOX (score: 11.5–11.9). FOX, specifically FOX-F, has been found to have a role in tooth formation [51]. Further investigations are needed to confirm the role that BMP2K might have in enamel hypoplasia.
Similar to the COHRA1 cohort, some of the top associated SNPs (rs11232439 and rs57555457) were intronic to genes (LOC100506433, MIR4300) with unknown biological functions.
A suggestive variant, rs62575356 (p = 2.7 × 10−7; Fig. 2g), is intronic to long intergenic non-protein coding RNA 474 (LOC101928775). This SNP is also located approximately 400 kb upstream of PAPPA, which has a role in bone formation, inflammation, and wound healing. In transcriptomic analysis of developing murine teeth, Pappa were found to be expressed in incisor teeth. In addition, Pappa was found to be in one of the networks genes that was centered in the NFκB pathway, which is one of the signaling pathways that regulate odontogenesis [52]. This SNP shows enhancer chromatin marks in many different cell types, including osteoblasts, which can be found in alveolar bone in the oral cavity, and this confirms the role that PAPPA plays during tooth development. However, it is still unclear how PAPPA could influence enamel hypoplasia, which suggests the need for more investigations.
Another suggestive variant, rs754479 (p = 8.0 × 10−7; Fig. 2h), is located 50 kb upstream of GSC. The encoded protein of GSC plays a role as a transcription factor. Deletion of Gsc in mice leads to many craniofacial defects. Gsc is detected in mice in the osteogenic mesenchyme of the mandible, which indicates its role in craniofacial development [53, 54].
Dental SCORE GWAS
No SNPs reached genome-wide significance; however, there were several regions that showed suggestive evidence. The top 4 lead SNPs were near genes with no biological relevance to tooth development, enamel, and/or craniofacial development. For example, rs2142491 (p = 6.18 × 10−8), is located approximately 300 kb from USP25, and this gene has been linked to Down syndrome [55]. Rs28504363 (p = 1.94 × 10−7) is approximately 100 kb downstream of ATOHA1, which has a role in neurogenesis [56]. rs36010081 is approximately 50 kb upstream of PPP2R5A (p = 2.67 × 10−7), and the protein encoded by this gene has been found to be expressed in skeletal muscles [57]. rs7780210 (p = 3.06 × 10−7) is intronic to CLEC2L. The biological function of this gene is still not fully understood. Further investigations can help in establishing if variation within these genomic regions may influence tooth development and/or enamel hypoplasia.
One of the suggestive variants in Dental SCORE was rs3010205 (p = 7.22 × 10−7; Fig. 2i), which is located approximately 100 kb from UBIAD1, which has a role in cholesterol metabolism [58]. rs3010205 is also located approximately 200 kb upstream of MTOR. The encoded protein of MTOR (mammalian target of rapamycin; mTOR), is a conserved protein kinase and required for multiple developmental processes, including differentiation, apoptosis, metabolism, and the development of neural cranial crest cells, which makes mTOR essential for craniofacial development. Furthermore, when disrupted, mTOR can cause defective facial growth, including defects or malformation in teeth, tongue, and palate [59].
GWAS Meta-Analysis Results
Meta-analysis was conducted to combine the GWAS results obtained from the four cohorts (see Table 3 for the lead SNPs results). The heterogeneity statistics results for the meta-analysis are shown in online supplementary Table S3. In addition, the lead SNPs from each individual cohort were looked up in the meta-GWAS results and in the other cohorts (online suppl. Table S4).
Table 3.
Meta-GWAS results
| SNPa | Chr | A1/A2b | Nearby gene(s)c | Meta -pd | Direction | Results for the meta-GWAS SNPs in each individual cohort |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POFC |
COHRA1 |
COHRA2 |
Dental SCORE |
||||||||||
| P | MAF | P | MAF | P | MAF | P | MAF | ||||||
| rs1359694 | 9 | A/C | PTPRD | 5.86E–07 | –––– | 0.0245855 | 0.1223 | 0.000493 | 0.1809 | 0.220907 | 0.1825 | 0.002401 | 0.1825 |
| rs12830414 | 12 | C/T | NAV3 | 6.07E–07 | ++++ | 0.0561199 | 0.09428 | 0.006441 | 0.06867 | 0.208079 | 0.07289 | 4.41 E–05 | 0.07289 |
| rs682846 | 19 | C/G | MYH14 | 6.36E–07 | –––– | 0.0304989 | 0.3097 | 0.003379 | 0.4438 | 0.023739 | 0.4283 | 0.009207 | 0.4283 |
| rs56282801e | 12 | A/G | DHX37 | 1.00E–06 | –––– | 0.0004047 | 0.1984 | 0.001016 | 0.2045 | 0.143019 | 0.3195 | 0.135073 | 0.3195 |
| rs4649222e | 1 | A/G | KIAA1804 | 1.83E–06 | ++++ | 0.0450894 | 0.2032 | 0.003782 | 0.2803 | 0.003620 | 0.2879 | 0.083122 | 0.2879 |
| rs2840075 | 4 | C/T | SLC4A4, ENAM | 2.20E–06 | –––– | 7.86E–06 | 0.07779 | 0.098322 | 0.1058 | 0.053585 | 0.0984 | 0.156516 | 0.0984 |
| rs62196465e | 20 | A/G | MACROD2 | 2.80E–06 | –––– | 0.0116859 | 0.05604 | 0.244040 | 0.05776 | 4.20E–05 | 0.05242 | 0.112376 | 0.05242 |
| rs16968212e | 15 | C/T | SCAPER | 2.92E–06 | ++++ | 0.2694265 | 0.06341 | 0.016560 | 0.07246 | 0.014049 | 0.07978 | 0.000683 | 0.07978 |
| rs9846530d | 3 | C/G | SEMA5B | 3.69E–06 | –––– | 0.1448401 | 0.2127 | 0.005046 | 0.2219 | 2.13E–05 | 0.2238 | 0.457281 | 0.2238 |
| rs7921002 | 10 | G/T | ZMIZ1 | 4.10E–06 | –––– | 0.0093433 | 0.3218 | 0.458316 | 0.178 | 0.002012 | 0.2056 | 0.005373 | 0.2056 |
All the SNPs presented in the table were imputed.
1 A1 is the effect allele, A2 is the other allele.2 Gene within the +500 kb window.3 There is no statistically significant heterogeneity in the meta-analysis results across the cohorts, see supplementary materials, Table S3.4 These SNPs were intronic.
None of the variants in the meta-analysis reached the genome-wide significance (p < 5 × 10−8); however, there were several that showed suggestive significance (5 × 10−8 < p < 5 × 10−6). The strongest suggestive signal was near rs1359694 (p = 5.86 × 10−7; Fig. 2j), which is intronic to PTPRD, and this gene encodes a member of the PTP (protein tyrosine phosphatase) family and has a role as a signaling molecule regulating different cellular processes such as differentiation, cell growth, and the mitotic cycle. PTPRD has been previously reported to be associated with smooth and pit-and-fissure surface caries in the primary dentition in children [60].
Several association signals were detected in the meta-analysis near genes that did not have clear biological relevance to tooth development or enamel hypoplasia. For example, rs12830414 (p = 6.07 × 10−7), is located approximately 400 kb from Neuron Navigator 3 (NAV3), which is highly expressed in the brain and the neuron system [61]; rs682846 (p = 6.36 × 10−7) is located within myosin heavy chain 14 (MYH14) which has been linked to deafness [62]; rs56282801 (p = 1.00 × 10−6) is located within DHX37 which has been linked to neurodevelopmental disorders [63]; and rs4649222 (p = 1.00 × 10−6) is intronic to KIAA1804, also known as MLK4, which can regulate the activation of transcription factor NF-κB [64].
One of the suggestive association signals in the meta-analysis was detected at rs2840075 (p = 2.20 × 10−6; Fig. 2k), which is located approximately 100 kb upstream of SLC4A4, and this gene encodes the sodium bicarbonate cotransporter (NBC), which has three main variants, the N-terminally spliced (NBCe1-A and NBCe1-B) and the C-terminally spliced (NBCe1-C). The NBCe1 is essential in regulating the pH of enamel matrix during tooth development, and more than one study, in humans and mice, has found that mutation in SLC4A4 can cause an abnormal enamel phenotype [65, 66, 67]. In addition, this SNP is a strong eQTL for SLC4A4. The lead SNP (rs2840075) and other SNPs (rs114654867, rs11729023) in high LD (r2> 0.8) at this locus show promoter and enhancer chromatin in many different cell types including osteoblasts. Motif analysis showed that the A allele of rs2840075 could change the affinity of Myc_disc5 (score 10.2–13.4). The MYC family, which Myc_disc5 belongs to, contains three members (C-MYC, N-MYC, and L-MYC). It has been reported that C-MYC specifically was found to be involved in tooth development [68]. All of this evidence could indicate that variation within this genomic region can influence tooth development and/or enamel hypoplasia.
Another suggestive variant from the meta-analysis results is rs62196465 (p = 2.6 × 10−6; Fig. 2m), which is intronic to MACROD2. In situ hybridization of mice showed that Macrod2 was significantly expressed in different tissues and organs, including teeth, during mouse embryonic development [69]. MACROD2 needs to be investigated in both animal and human studies to understand the exact role it might have during tooth development.
Functional Annotation of Meta-GWAS Results
FUMA of the meta-GWAS (Table 3) was used to obtain more information regarding the different biological aspects of the observed associations. FUMA meta-GWAS mapped a total of 20 genes to the 18 risk loci identified (online suppl. Fig. 1). These genes' expression was examined across different human tissues from different databases. FUMA results of the meta-GWAS risk loci (p < 5.0 × 10−6) are shown in online supplementary Figure 1. In addition, functional mapping linked lead SNPs and nearby SNPs in tight LD with the GWAS catalog (https://www.ebi.ac.uk/gwas/). All of the results of the GWAS catalog-based annotation are presented in online supplementary Table S5.
The gene-based test based on the meta-GWAS results of all cohorts was computed by MAGMA (multi-marker analysis of genomic annotation) using the default settings implemented in FUMA. Lead SNPs from the meta-GWAS analysis were mapped to 19,182 protein coding genes. The genome-wide significance level was calculated based on the number of tested genes and it was set at p of 0.05/19,182 = 2.61 × 10−6. None of the genes reached the genome-wide significance level. A Manhattan plot of the gene-based test are shown in online supplementary Figure 2.
Gene set enrichment analyses were conducted with GENE2FUNC implemented in FUMA. The Bonferroni-corrected threshold for significant gene set enrichment was set at p ≤ 0.05. A gene set at chr17q23 was overexpressed (p = 2.46 × 10−7) in the positional gene sets (MSigDB C1), which is a set of genes that can help in identifying effects based on chromosomal changes (e.g., deletions or amplifications). Full results of the gene set enrichment analyses are shown in online supplementary Table S6.
Discussion
Enamel hypoplasia is a multifactorial condition believed to be caused by the interaction between environmental and genetic factors. Some epidemiological studies show that the prevalence of enamel hypoplasia differs depending on several factors, including which teeth are considered, the examination method, and the population investigated. The reported prevalence of enamel hypoplasia in the current study was 7.1% combining across cohorts, falling within the reported range of enamel hypoplasia prevalence in other published studies (0.8 to 17% for the permanent dentition) [9, 10, 11, 12]. However, there was considerable variation in prevalence across our 4 study cohorts, which may be due to factors differing across the cohorts such as population diversity, environmental, and/or socioeconomic factors, and enamel hypoplasia assessment methods.
In our current study we found that the most affected teeth by enamel hypoplasia in POFC were the maxillary left central incisors (7.5%), followed by maxillary right central incisors (7.33%) and upper right lateral incisors (7.33%). In previous studies [10, 12], maxillary incisors were found to be the most affected teeth by enamel hypoplasia. In COHRA1, the most affected teeth with enamel hypoplasia were the maxillary right central incisors (7.84%), followed by the maxillary right first molars (6.86%) and maxillary left lateral incisors (5.88%). In COHRA2, the most affected teeth by enamel hypoplasia were maxillary right first molar (6.60%), followed by maxillary left first molar (6.13%) and maxillary left lateral incisor (5.18%). Finally, the most affected teeth with enamel hypoplasia in Dental SCORE were the maxillary right first molar (11.34%), followed by maxillary right second molar (10.31%) and maxillary left lateral incisors (6.18%). These results are shown in Table 1. Previous studies found that incisors, especially in the maxilla, and molars are the most affected teeth with enamel hypoplasia [10, 12], and our findings were similar to what has been found in those studies.
The cohorts included in this study had somewhat different protocols for collecting and measuring enamel hypoplasia from the participants. Thus, to harmonize analyses across the different cohorts, we used a binary definition of enamel hypoplasia, i.e., approximating a case-control study design that can provide a threshold level of an underlying continuum of enamel hypoplasia risk. Similar binary traits were also successfully used in the past in studying other complex oral traits, such as dental caries [70]. Note that recording enamel hypoplasia as a binary trait did not contain information on its extent or severity, which may have reduced the statistical power of our analysis. Thus, ideally, future studies should capture the full extent of the trait. However, it is important to emphasize that although we had varying protocols to collect enamel hypoplasia in our study, variability in phenotype definition should not lead to biased genetic results, although it may have reduced statistical power [71].
To the best of our knowledge, this study represents the first large-scale GWAS performed for enamel hypoplasia. The main aim of this study was to identify genetic loci that are associated with enamel hypoplasia. We were able to identify and nominate several variants near genes with reasonable biological roles in tooth and/or enamel development.
The only previously published GWAS in the literature that focused on enamel defects was on molar-incisor hypomineralization (MIH) in children from Munich [72]. MIH differs from the enamel hypoplasia trait of the current study, but it can be hard to distinguish teeth that are affected with enamel hypoplasia from teeth affected with MIH. Enamel hypoplasia is characterized by reduced thickness of enamel, while MIH affects the translucency of enamel [73]. The top association signal with MIH (rs13058467, p = 3.72 × 10−7) was near SCUBE1, which plays a role in the development of the tooth and in craniofacial development [72, 74]. In our meta-analysis the p value for association with enamel hypoplasia for rs13058467 was 0.328. We also looked in the region around that lead SNP (+500 kb), but no other SNPs in that region were significantly associated in our meta-analysis results.
The current study identified several genome-wide significant and suggestive association signals, near genes that have plausible roles, directly or indirectly, in tooth development and enamel, and thus could possibly contribute to increasing the risk of developing enamel hypoplasia. These include but are not limited to: BMP2K, which is involved in tooth development [46]; MTOR, which has been implicated in craniofacial development and the differentiation of odontoblasts [75]; MACROD2, which has been reported to be expressed in teeth during embryonic development in mice [69]; SLC4A4, which has strong biological evidence for the role it plays in influencing enamel hypoplasia, and PTPRD, which has been reported to be associated with smooth and pit-and-fissure surface caries in the primary dentition in children [60].
The fact that PTPRD has also been found to be associated with dental caries indicates that there may be an etiologic relationship between dental caries and enamel hypoplasia [60]. Indeed, enamel hypoplasia can create an enhanced environment for adherence and retention of cariogenic bacteria for a long period of time, which can increase the risk of developing dental caries [8] and can be considered as a potential confounder for dental caries. In our different cohorts, we had an average mean score of 6.88 of dental caries assessed by counts of decayed and filled teeth due to caries (DFT index), which is considered to be high. The highest DFT mean score was seen in Dental SCORE (12.07), which was not surprising given that most participants in Dental SCORE were older, on average, than the other cohorts' participants. We used the DFT index instead of DMFT (counts of decayed, missing, and filled teeth due to caries) because we had incomplete information regarding the underlying cause of missing teeth, i.e., whether teeth were missing due to caries or for other reasons, such as injury.
The fact that some of the prioritized loci are related to pathways that regulate the tooth development process is consistent with the current understanding of the etiology of dental anomalies, which include enamel hypoplasia. For example, one of the lead SNPs in the POFC GWAS was near ULK4, which has an important role in regulating the wnt signaling pathway. Thus, mutations in wnt signaling components could lead to developing dental anomalies, such as tooth agenesis [31]. In addition, enamel hypoplasia was one of the manifestations that some patients reported as a result of a rare deletion of ULK4 [32]. This evidence suggests that ULK4 has a possible role during odontogenesis (tooth development) and/or enamel hypoplasia.
Under the hypothesis that enamel development genes could influence enamel hypoplasia, we also reviewed the p values of 23 SNPs in the meta-analysis results that have been previously examined and reported [76, 77, 78] in and near some of the enamel matrix candidate genes (ENAM, AMELX, AMBN, TFIP11, TUFT1). In addition, we also investigated genes that were recently reviewed [79] to be involved in hereditary enamel defects in humans. Therefore, we investigated a total of 32 SNPs. A threshold of a p value less than 0.001 was chosen to declare statistical significance, which corresponds to the Bonferroni correction for 32 SNPs. Interestingly, two SNPs showed statistical significance (p < 0.001): rs10518733 within WDR72, and rs11136305 within FAM83H. The full list of the results of these 32 variants is presented in Table 4.
Table 4.
Previously identified SNPs in enamel matrix genes (ENAM, TUFT1, AMBN, TFIP11, and AMELX)
| SNP | Gene | Chr | BP | A1/A2a | p | Type | dbSNP functional annotation |
|---|---|---|---|---|---|---|---|
| rs17733915 | AMBN | 4 | 71447093 | T/C | 0.1846 | Genotyped | Upstream |
| rs4694075 | AMBN | 4 | 71466914 | T/C | 0.4764 | Imputed | Intronic |
| rs17149026 | AMBN | 4 | 71440940 | G/T | 0.8275 | Imputed | Upstream |
| rs2106416 | AMLEX | 23 | 11316742 | T/C | 0.4535 | Imputed | Synonymous |
| rs946252 | AMLEX | 23 | 11313027 | T/C | 0.7673 | Imputed | Intronic |
| rs1967376 | ENAM | 4 | 71501744 | C/T | 0.3458 | Imputed | Intronic |
| rs12640848 | ENAM | 4 | 71506412 | G/A | 0.6669 | Imputed | Intronic |
| rs2097470 | TFIP11 | 22 | 26904965 | T/C | 0.1351 | Imputed | Intronic |
| rs713900 | TFIP11 | 22 | 26898242 | A/G | 0.214 | Imputed | Intronic |
| rs134135 | TFIP11 | 22 | 26898962 | C/G | 0.5798 | Imputed | Intronic |
| rs5997096 | TFIP11 | 22 | 26895957 | C/T | 0.6212 | Imputed | Intronic |
| rs134136 | TFIP11 | 22 | 26899474 | T/C | 0.6672 | Imputed | Intronic |
| rs134134 | TFIP11 | 22 | 26898891 | T/C | 0.7116 | Imputed | Intronic |
| rs134145 | TFIP11 | 22 | 26909750 | A/G | 0.7835 | Genotyped | Intronic |
| rs6005060 | TFIP11 | 22 | 26895736 | A/T | 0.9559 | Imputed | Intronic |
| rs2337359 | TUFT1 | 1 | 151495796 | C/T | 0.004234 | Imputed | Upstream |
| rs10158855 | TUFT1 | 1 | 151515654 | G/T | 0.07768 | Imputed | Intronic |
| rs17640579 | TUFT1 | 1 | 151521933 | G/A | 0.1621 | Imputed | Intronic |
| rs3790506 | TUFT1 | 1 | 151538366 | A/G | 0.198 | Imputed | Intronic |
| rs16833391 | TUFT1 | 1 | 151547253 | T/C | 0.52 | Imputed | Intronic |
| rs1045298 | TUFT1 | 1 | 151510825 | T/C | 0.5601 | Imputed | 3' UTR |
| rs12749 | TUFT1 | 1 | 151555741 | A/G | 0.8748 | Imputed | 3' UTR |
| rs4970957 | TUFT1 | 1 | 151517388 | G/A | 0.9746 | Imputed | Intronic |
| rs10518733 | WDR72 | 15 | 53940307 | A/C | 0.0005654 | Genotyped | Intronic |
| rs11136305 | FAM83H | 8 | 144689914 | C/T | 0.001001 | Imputed | Upstream |
| rs3745542 | KLK4 | 9 | 51587643 | C/T | 0.002304 | Genotyped | 140 bp 5' of KLK14 |
| rs8005614 | SLC24A4 | 14 | 92971222 | G/T | 0.002784 | Imputed | 8.6 kb 3' of SLC24A4 |
| rs2819865 | RUNX2 | 6 | 45443176 | A/G | 0.003116 | Imputed | Intronic |
| rs12804929 | MMP20 | 11 | 10264250 | G/A | 0.003788 | Imputed | Downstream |
| rs3803878 | DLX3 | 17 | 48067581 | G/C | 0.00385 | Imputed | 3' UTR |
| rs10044464 | ADAMTS2 | 5 | 178692688 | C/T | 0.004017 | Imputed | Intronic |
| rs9895891 | FAM20A | 17 | 66329586 | G/A | 0.004428 | Imputed | Downstream |
A1 is the effect allele, A2 is the other allele.
Additional SNPs near the enamel matrix candidate genes (ENAM, AMELX, AMBN, TFIP11, TUFT1) emerged within the 500 kb regions flanking these genes in our meta-analysis results. A total of 8,544 SNPs were in and near these enamel genes and considering that some of these SNPs are in LD, the Li and Ji method for determining the number of independent tests [80] was used leading to an adjusted threshold p value of 1.79 × 10−4 to declare statistical significance. There was one SNP that reached statistical significance (rs2840075, p = 2.20 × 10−6), located approximately 500 kb downstream from ENAM. The encoded protein of ENAM (enamelin) is required for the proper development of teeth and enamel [4]. AMBN, another gene that has a role in the development of enamel, is located approximately 600 kb upstream of this SNP, although given the distance of this SNP from AMBN (600 kb) it is unclear if this SNP affects AMBN function or expression. The full list of the association results with the enamel matrix candidate genes is shown in online supplementary Table S7.
One of the challenges in this study was the lack of replication of association signals across cohorts. This could be due to several possible sources of heterogeneity, such as different environmental factors, different phenotype assessment and study designs, different ethnic groups, and using different genotyping chips. In addition, the SNPs that are in linkage disequilibrium with the lead SNPs may differ from one ethnic group to another; formal assessment of the heterogeneity in genetic effects (I2) showed little evidence for heterogeneity in the top associated loci, but with only four cohorts the power to detect heterogeneity may have been limited.
There are limitations of the current study. First, the study only identified the associations between genetic variants and enamel hypoplasia, not causation or the biological effect of these variants and implicated genes on enamel hypoplasia. Second, the p values from EMMAX are accurate but effect sizes are not informative because EMMAX is based on mixed models rather than a generalized mixed model. Finally, the study did not replicate most of the association signals across the different cohorts. These limitations are balanced by the fact that several of the association signals described and investigated in our study were near genes supported by a body of literature with involvement in oral and tooth biological processes.
The current study provides a wealth of new information on possible candidate genes and loci that might be involved in enamel hypoplasia and will require additional investigation in animal and human studies, replication, and validation in larger studies. In addition, other suggestive signals near genes with no previously known biological roles in tooth development or enamel hypoplasia should also be further investigated.
This study is the first GWAS that aimed to identify genetic loci that associated with enamel hypoplasia and nominated several for further investigation, including ULK4, CCK, ADAR1, PYGO1, CELSR1, BMP2K, MTOR, PAPPA, and SLC4A4 because of their plausible biological roles in enamel development. Further studies are required to confirm the role these genes play in enamel development during odontogenesis using both human and mouse models. In addition, more research to replicate the analyses in independent cohorts is essential.
Statement of Ethics
Study approval was granted by the University of Pittsburgh Institutional Review Board (coordinating center approval No. CR19030367-003, Pittsburgh site approval No. CR19080127-00). All participants provided written informed consent for themselves and for children younger than 18 years, informed consent was obtained from their parents or their legally authorized representative. Local ethical approval was obtained at each site and all methods in this study were performed in accordance with the Institutional Review Board policies and guidelines of the University of Pittsburgh and all of the other sites. See also online supplementary Table S2.
Conflict of Interest Statement
The authors declare no potential conflicts of interest with respect to the publication of this article.
Funding Sources
This work was funded through grants from the National Institutes of Health (NIH) including: R01-DE016148 (M.L.M, S.M.W), X01-HG007485 (M.L.M), R01-DE014899 (M.L.M, D.W.M), U01-DE018903 (M.L.M), X01-HG009878 (J.R.S), R21-DE016930 (M.L.M), R03-DE024264 (J.R.S), R01-DE015667 (L.M.M.U), K99-DE024571 (C.J.B), R25-MD007607 (C.J.B), U54-MD007587 (C.J.B), R01-DD000295 (G.L.W), U01-DE024425 (M.L.M). Genotyping and data cleaning were done by the Center for Inherited Disease Research, through NIH contracts HHSN268200782096C and HHSN268201200008I.
Author Contributions
R.N.A. generated the data set for the analyses. R.N.A. analyzed data. R.N.A., M.L.M., J.R.S., S.M.W. defined the outcomes to be studied. R.N.A. wrote the first draft of the manuscript. M.L.M, J.R.S, S.M.W, B.J.H, L.M.M.U, and K.N. critically reviewed the manuscript. R.N.A. and M.L.M. designed the study. R.N.A. interpreted the data. M.L.M., C.S., F.W.B.D., K.N., C.P.R.M., C.V.-R., F.A.P., I.M.O., C.J.B., G.L.W., J.R.S., S.M.W., J.M., R.E.L., A.R.V., C.P., S.E.R., and L.M.M.U. collected and interpreted the data. R.N.A. and M.L.M. generated the final draft of the manuscript. All authors reviewed the final manuscript.
Data Availability Statement
The datasets analyzed in this paper are available in the dbGaP repository (Studies Accession: phs000774.v2.p1, phs001591.v1.p1, phs000095.v3.p1). See also online supplementary Table S2. Authorized members and NIH Investigators are eligible to apply to access these datasets.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgements
We would like to thank the participating families from around the world who made this study possible, and who also provided inspiration to the investigators and everyone involved in these studies. The tireless efforts by our beloved staff and collaborators from different sites were necessary for the success of this study.
Funding Statement
This work was funded through grants from the National Institutes of Health (NIH) including: R01-DE016148 (M.L.M, S.M.W), X01-HG007485 (M.L.M), R01-DE014899 (M.L.M, D.W.M), U01-DE018903 (M.L.M), X01-HG009878 (J.R.S), R21-DE016930 (M.L.M), R03-DE024264 (J.R.S), R01-DE015667 (L.M.M.U), K99-DE024571 (C.J.B), R25-MD007607 (C.J.B), U54-MD007587 (C.J.B), R01-DD000295 (G.L.W), U01-DE024425 (M.L.M). Genotyping and data cleaning were done by the Center for Inherited Disease Research, through NIH contracts HHSN268200782096C and HHSN268201200008I.
References
- 1.Thesleff I. The genetic basis of tooth development and dental defects. Am J Med Genet A. 2006 Dec 1;140((23)):2530–2535. doi: 10.1002/ajmg.a.31360. [DOI] [PubMed] [Google Scholar]
- 2.Bailleul-Forestier I, Molla M, Verloes A, Berdal A. The genetic basis of inherited anomalies of the teeth. Eur J Med Genet. 2008;51((4)):273–291. doi: 10.1016/j.ejmg.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Brook AH, Griffin RC, Townsend G, Levisianos Y, Russell J, Smith RN. Variability and patterning in permanent tooth size of four human ethnic groups. Arch Oral Biol. 2009 Dec;54 Suppl 1:S79–85. doi: 10.1016/j.archoralbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Gerreth K, Zaorska K, Zabel M, Nowicki M, Borysewicz-Lewicka M. Significance of genetic variations in developmental enamel defects of primary dentition in Polish children. Clin Oral Investig. 2018;22((1)):321–329. doi: 10.1007/s00784-017-2115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suckling GW. Developmental defects of enamel: historical and present-day perspectives of their pathogenesis. Adv Dent Res. 1989 Sep;3((2)):87–94. doi: 10.1177/08959374890030022901. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Navia JM, Bian JY. Caries experience in deciduous dentition of rural Chinese children 3–5 years old in relation to the presence or absence of enamel hypoplasia. Caries Res. 1996;30((1)):8–15. doi: 10.1159/000262130. [DOI] [PubMed] [Google Scholar]
- 7.Lai PY, Seow WK, Tudehope DI, Rogers Y. Enamel hypoplasia and dental caries in very-low birthweight children: a case-controlled, longitudinal study. Pediatr Dent. 1997 Jan-Feb;19((1)):42–49. [PubMed] [Google Scholar]
- 8.Hong L, Levy SM, Warren JJ, Broffitt B. Association between enamel hypoplasia and dental caries in primary second molars: a cohort study. Caries Res. 2009;43((5)):345–353. doi: 10.1159/000231571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lunardelli SE, Peres MA. Prevalence and distribution of developmental enamel defects in the primary dentition of pre-school children. Braz Oral Res. 2005 Apr-Jun;19((2)):144–149. doi: 10.1590/s1806-83242005000200013. [DOI] [PubMed] [Google Scholar]
- 10.Seow WK, Ford D, Kazoullis S, Newman B, Holcombe T. Comparison of enamel defects in the primary and permanent dentitions of children from a low-fluoride district in Australia. Pediatr Dent. 2011 May-Jun;33((3)):207–212. [PubMed] [Google Scholar]
- 11.Chauhan D, Chauhan T. Prevalence of developmental defects of enamel in mixed and permanent dentition of 9 and 12 year old children of Himachal Pradesh, India: a cross sectional study. Int J Health Allied Sci. 2013;2:185–188. [Google Scholar]
- 12.Robles MJ, Ruiz M, Bravo-Perez M, González E, Peñalver MA. Prevalence of enamel defects in primary and permanent teeth in a group of schoolchildren from Granada (Spain) Med Oral Patol Oral Cir Bucal. 2013;18((2)):e187–93. doi: 10.4317/medoral.18580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarkson J. Review of terminology, classifications, and indices of developmental defects of enamel. Adv Dent Res. 1989 Sep;3((2)):104–109. doi: 10.1177/08959374890030020601. [DOI] [PubMed] [Google Scholar]
- 14.Howe BJ, Cooper ME, Vieira AR, Weinberg SM, Resick JM, Nidey NL, et al. Spectrum of dental phenotypes in nonsyndromic orofacial clefting. J Dent Res. 2015;94((7)):905–912. doi: 10.1177/0022034515588281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polk DE, Weyant RJ, Crout RJ, McNeil DW, Tarter RE, Thomas JG, et al. Study protocol of the Center for Oral Health Research in Appalachia (COHRA) etiology study. BMC Oral Health. 2008;8:18. doi: 10.1186/1472-6831-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neiswanger K, McNeil DW, Foxman B, Govil M, Cooper ME, Weyant RJ, et al. Oral health in a sample of pregnant women from Northern Appalachia. Int J Dent. 2015;2015:469376. doi: 10.1155/2015/469376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiyer AN, Kip KE, Marroquin OC, Mulukutla SR, Edmundowicz D, Reis SE. Racial differences in coronary artery calcification are not attributed to differences in lipoprotein particle sizes: the heart strategies concentrating on risk evaluation (Heart SCORE) study. Am Heart J. 2007 Feb;153((2)):328–334. doi: 10.1016/j.ahj.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Shaffer JR, Zeng Z, Begum F, Vieira AR, Noel J, et al. Genome-wide association scan of dental caries in the permanent dentition. BMC Oral Health. 2012;12:57. doi: 10.1186/1472-6831-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet. 2016 Jul 1;25((13)):2862–2872. doi: 10.1093/hmg/ddw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009 Jun;5((6)):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov 1;491((7422)):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shungin D, Haworth S, Divaris K, Agler CS, Kamatani Y, Keun Lee M, et al. Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat Commun. 2019;10((1)):2773–2773. doi: 10.1038/s41467-019-10630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010 Sep;34((6)):591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett SN, Caporaso N, Fitzpatrick AL, Agrawal A, Barnes K, Boyd HA, et al. Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience. Genet Epidemiol. 2011;35((3)):159–173. doi: 10.1002/gepi.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods. 2014;11((4)):407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42((4)):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26((17)):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012 Jan;40((Database issue)):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017 Nov;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010 Sep 15;26((18)):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura M, Nemoto E. Role of the Wnt signaling molecules in the tooth. Jpn Dent Sci Rev. 2016;52((4)):75–83. doi: 10.1016/j.jdsr.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M, Guan Z, Shen Q, Flinter F, Domínguez L, Ahn JW, et al. Ulk4 Regulates neural stem cell pool. Stem Cells. 2016 Sep;34((9)):2318–2331. doi: 10.1002/stem.2423. [DOI] [PubMed] [Google Scholar]
- 33.Liu M, Xu P, O'Brien T, Shen S. Multiple roles of Ulk4 in neurogenesis and brain function. Neurogenesis. 2017;4((1)):e1313646. doi: 10.1080/23262133.2017.1313646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nurbaeva MK, Eckstein M, Devotta A, Saint-Jeannet JP, Yule DI, Hubbard MJ, et al. Evidence that calcium entry into calcium-transporting dental enamel cells is regulated by cholecystokinin, acetylcholine and ATP. Front Physiol. 2018;9:801. doi: 10.3389/fphys.2018.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacruz RS, Smith CE, Bringas P, Chen YB, Smith SM, Snead ML, et al. Identification of novel candidate genes involved in mineralization of dental enamel by genome-wide transcript profiling. J Cell Physiol. 2012;227((5)):2264–2275. doi: 10.1002/jcp.22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kantaputra PN, Chinadet W, Ohazama A, Kono M. Dyschromatosis symmetrica hereditaria with long hair on the forearms, hypo/hyperpigmented hair, and dental anomalies: report of a novel ADAR1 mutation. Am J Med Genet A. 2012;158A((9)):2258–2265. doi: 10.1002/ajmg.a.35488. [DOI] [PubMed] [Google Scholar]
- 37.Cantu C, Pagella P, Shajiei TD, Zimmerli D, Valenta T, Hausmann G, et al. A cytoplasmic role of Wnt/beta-catenin transcriptional cofactors Bcl9, Bcl9l, and Pygopus in tooth enamel formation. Sci Signal. 2017 Feb 7;10((465)):eaah4598. doi: 10.1126/scisignal.aah4598. [DOI] [PubMed] [Google Scholar]
- 38.Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, et al. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors . Dev Cell. 2012 Aug 14;23((2)):317–328. doi: 10.1016/j.devcel.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juuri E, Jussila M, Seidel K, Holmes S, Wu P, Richman J, et al. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development. 2013 Apr;140((7)):1424–1432. doi: 10.1242/dev.089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obara N, Suzuki Y, Irie K, Shibata S. Expression of planar cell polarity genes during mouse tooth development. Arch Oral Biol. 2017;83((83)):85–91. doi: 10.1016/j.archoralbio.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 41.van den Oord EJCG, Kuo PH, Hartmann AM, Webb BT, Möller HJ, Hettema JM, et al. Genomewide association analysis followed by a replication study implicates a novel candidate gene for neuroticism. Arch Gen Psychiatr. 2008;65((9)):1062–1071. doi: 10.1001/archpsyc.65.9.1062. [DOI] [PubMed] [Google Scholar]
- 42.Taira TM, Lima V, Prado DS, Silva TA, Issa JPM, da Silva LAB, et al. NLRP12 Attenuates inflammatory bone loss in experimental apical periodontitis. J Dent Res. 2019 Apr;98((4)):476–484. doi: 10.1177/0022034518820289. [DOI] [PubMed] [Google Scholar]
- 43.Wang K, Li J. Overexpression of ANXA3 is an independent prognostic indicator in gastric cancer and its depletion suppresses cell proliferation and tumor growth. Oncotarget. 2016;7((52)):86972–84. doi: 10.18632/oncotarget.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishidou Y, Kitajima I, Obama H, Maruyama I, Murata F, Imamura T, et al. Enhanced expression of type I receptors for bone morphogenetic proteins during bone formation. J Bone Miner Res. 1995 Nov;10((11)):1651–1659. doi: 10.1002/jbmr.5650101107. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura Y, Wakitani S, Nakayama J, Wakabayashi S, Horiuchi H, Takaoka K. Temporal and spatial expression profiles of BMP receptors and noggin during BMP-2-induced ectopic bone formation. J Bone Miner Res. 2003 Oct;18((10)):1854–1862. doi: 10.1359/jbmr.2003.18.10.1854. [DOI] [PubMed] [Google Scholar]
- 46.Kearns AE, Donohue MM, Sanyal B, Demay MB. Cloning and characterization of a novel protein kinase that impairs osteoblast differentiation in vitro. J Biol Chem. 2001 Nov 9;276((45)):42213–42218. doi: 10.1074/jbc.M106163200. [DOI] [PubMed] [Google Scholar]
- 47.Kanzler B, Foreman RK, Labosky PA, Mallo M. BMP signaling is essential for development of skeletogenic and neurogenic cranial neural crest. Development. 2000 Mar;127((5)):1095–1104. doi: 10.1242/dev.127.5.1095. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Cao X. BMP signaling and skeletogenesis. Ann NY Acad Sci. 2006;1068:26–40. doi: 10.1196/annals.1346.006. [DOI] [PubMed] [Google Scholar]
- 49.Rakian A, Yang WC, Gluhak-Heinrich J, Cui Y, Harris MA, Villarreal D, et al. Bone morphogenetic protein-2 gene controls tooth root development in coordination with formation of the periodontium. Int J Oral Sci. 2013;5((2)):75–84. doi: 10.1038/ijos.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Y, Qian Y, Zhang J, Gong M, Wang Y, Gu N, et al. Genetic variants of bmp2 and their association with the risk of non-syndromic tooth agenesis. PLoS One. 2016;11((6)):e0158273. doi: 10.1371/journal.pone.0158273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu P, Balczerski B, Ciozda A, Louie K, Oralova V, Huysseune A, et al. Fox proteins are modular competency factors for facial cartilage and tooth specification. Development. 2018;145((12)):dev165498. doi: 10.1242/dev.165498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laugel-Haushalter V, Paschaki M, Thibault-Carpentier C, Dembelé D, Dollé P, Bloch-Zupan A. Molars and incisors: show your microarray IDs. BMC Res Notes. 2013 Mar 26;6:113. doi: 10.1186/1756-0500-6-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivera-Perez JA, Wakamiya M, Behringer RR. Goosecoid acts cell autonomously in mesenchyme-derived tissues during craniofacial development. Development. 1999 Sep;126((17)):3811–3821. doi: 10.1242/dev.126.17.3811. [DOI] [PubMed] [Google Scholar]
- 54.Ganeshkar SV, Rai AK, Rozario JE. The master of craniofacial orchestra: homeobox genes and neural crest cells. Intl Arch Integrat Med. 2015;2((7)):162–170. [Google Scholar]
- 55.Valero R, Bayés M, Francisca Sánchez-Font M, González-Angulo O, Gonzàlez-Duarte R, Marfany G. Characterization of alternatively spliced products and tissue-specific isoforms of USP28 and USP25. Genome Biol. 2001;2((10)):RESEARCH0043. doi: 10.1186/gb-2001-2-10-research0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mulvaney J, Dabdoub A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J Assoc Res Otolaryngol. 2012;13((3)):281–293. doi: 10.1007/s10162-012-0317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCright B, Brothman AR, Virshup DM. Assignment of human protein phosphatase 2A regulatory subunit genes b56alpha, b56beta, b56gamma, b56delta, and b56epsilon (PPP2R5A-PPP2R5E), highly expressed in muscle and brain, to chromosome regions 1q41, 11q12, 3p21, 6p21.1. Genomics. 1996 Aug 15;36((1)):168–170. doi: 10.1006/geno.1996.0438. [DOI] [PubMed] [Google Scholar]
- 58.Morales CR, Grigoryeva LS, Pan X, Bruno L, Hickson G, Ngo MH, et al. Mitochondrial damage and cholesterol storage in human hepatocellular carcinoma cells with silencing of UBIAD1 gene expression. Mol Genet Metab Rep. 2014;1:407–411. doi: 10.1016/j.ymgmr.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nie X, Zheng J, Ricupero CL, He L, Jiao K, Mao JJ. mTOR acts as a pivotal signaling hub for neural crest cells during craniofacial development. PLoS Genet. 2018 Jul;14((7)):e1007491. doi: 10.1371/journal.pgen.1007491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng Z, Feingold E, Wang X, Weeks DE, Lee M, Cuenco KT, et al. Genome-wide association study of primary dentition pit-and-fissure and smooth surface caries. Caries Res. 2014;48((4)):330–338. doi: 10.1159/000356299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maes T, Barceló A, Buesa C. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from caenorhabditis elegans. Genomics. 2002;80((1)):21–30. doi: 10.1006/geno.2002.6799. [DOI] [PubMed] [Google Scholar]
- 62.Kim BJ, Kim AR, Han JH, Lee C, Oh DY, Choi BY. Discovery of MYH14 as an important and unique deafness gene causing prelingually severe autosomal dominant nonsyndromic hearing loss. J Gene Med. 2017 Apr;19((4)):2950. doi: 10.1002/jgm.2950. [DOI] [PubMed] [Google Scholar]
- 63.Paine I, Posey JE, Grochowski CM, Jhangiani SN, Rosenheck S, Kleyner R, et al. Paralog studies augment gene discovery: DDX and DHX genes. Am J Hum Genet. 2019 Aug 1;105((2)):302–316. doi: 10.1016/j.ajhg.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim SH, Ezhilarasan R, Phillips E, Gallego-Perez D, Sparks A, Taylor D, et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-κB-dependent manner. Cancer Cell. 2016 Feb 8;29((2)):201–213. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lacruz RS, Smith CE, Moffatt P, Chang EH, Bromage TG, Bringas P, Jr, et al. Requirements for ion and solute transport, and pH regulation during enamel maturation. J Cell Physiol. 2012 Apr;227((4)):1776–1785. doi: 10.1002/jcp.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jalali R, Guo J, Zandieh-Doulabi B, Bervoets TJ, Paine ML, Boron WF, et al. NBCe1 (SLC4A4) a potential pH regulator in enamel organ cells during enamel development in the mouse. Cell Tissue Res. 2014 Nov;358((2)):433–442. doi: 10.1007/s00441-014-1935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wen X, Kurtz I, Paine ML. Prevention of the disrupted enamel phenotype in Slc4a4-null mice using explant organ culture maintained in a living host kidney capsule. PLoS One. 2014;9((5)):e97318. doi: 10.1371/journal.pone.0097318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peng Z, Liu L, Wei X, Ling J. Expression of Oct-4, SOX-2, and MYC in dental papilla cells and dental follicle cells during in-vivo tooth development and in-vitro co-culture. Eur J Oral Sci. 2014 Aug;122((4)):251–258. doi: 10.1111/eos.12141. [DOI] [PubMed] [Google Scholar]
- 69.Maas NM, Van de Putte T, Melotte C, Francis A, Schrander-Stumpel CT, Sanlaville D, et al. The C20orf133 gene is disrupted in a patient with Kabuki syndrome. J Med Genet. 2007 Sep;44((9)):562–569. doi: 10.1136/jmg.2007.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Opal S, Garg S, Jain J, Walia I. Genetic factors affecting dental caries risk. Aust Dent J. 2015 Mar;60((1)):2–11. doi: 10.1111/adj.12262. [DOI] [PubMed] [Google Scholar]
- 71.van der Sluis S, Verhage M, Posthuma D, Dolan CV. Phenotypic complexity, measurement bias, and poor phenotypic resolution contribute to the missing heritability problem in genetic association studies. PLoS One. 2010 Nov 10;5((11)):e13929. doi: 10.1371/journal.pone.0013929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuhnisch J, Thiering E, Heitmuller D, Tiesler CM, Grallert H, Heinrich-Weltzien R, et al. Genome-wide association study (GWAS) for molar-incisor hypomineralization (MIH) Clin Oral Investig. 2014;18((2)):677–682. doi: 10.1007/s00784-013-1054-8. [DOI] [PubMed] [Google Scholar]
- 73.Garg N, Jain AK, Saha S, Singh J. Essentiality of early diagnosis of molar incisor hypomineralization in children and review of its clinical presentation, etiology and management. Int J Clin Pediatr Dent. 2012;5((3)):190–196. doi: 10.5005/jp-journals-10005-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xavier GM, Sharpe PT, Cobourne MT. Scube1 is expressed during facial development in the mouse. J Exp Zool B Mol Dev Evol. 2009 Jul 15;312B((5)):518–524. doi: 10.1002/jez.b.21260. [DOI] [PubMed] [Google Scholar]
- 75.Kim JK, Baker J, Nor JE, Hill EE. mTor plays an important role in odontoblast differentiation. J Endod. 2011 Aug;37((8)):1081–1085. doi: 10.1016/j.joen.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 76.Jeremias F, Koruyucu M, Küchler EC, Bayram M, Tuna EB, Deeley K, et al. Genes expressed in dental enamel development are associated with molar-incisor hypomineralization. Arch Oral Biol. 2013;58((10)):1434–1442. doi: 10.1016/j.archoralbio.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaffer JR, Carlson JC, Stanley BO, Feingold E, Cooper M, Vanyukov MM, et al. Effects of enamel matrix genes on dental caries are moderated by fluoride exposures. Hum Genet. 2015 Feb;134((2)):159–167. doi: 10.1007/s00439-014-1504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piekoszewska-Zietek P, Turska-Szybka A, Olczak-Kowalczyk D. Single nucleotide polymorphism in the aetiology of caries: systematic literature review. Caries Res. 2017;51((4)):425–435. doi: 10.1159/000476075. [DOI] [PubMed] [Google Scholar]
- 79.Wright JT, Carrion IA, Morris C. The molecular basis of hereditary enamel defects in humans. J Dent Res. 2015 Jan;94((1)):52–61. doi: 10.1177/0022034514556708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005 Sep;95((3)):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Data Availability Statement
The datasets analyzed in this paper are available in the dbGaP repository (Studies Accession: phs000774.v2.p1, phs001591.v1.p1, phs000095.v3.p1). See also online supplementary Table S2. Authorized members and NIH Investigators are eligible to apply to access these datasets.