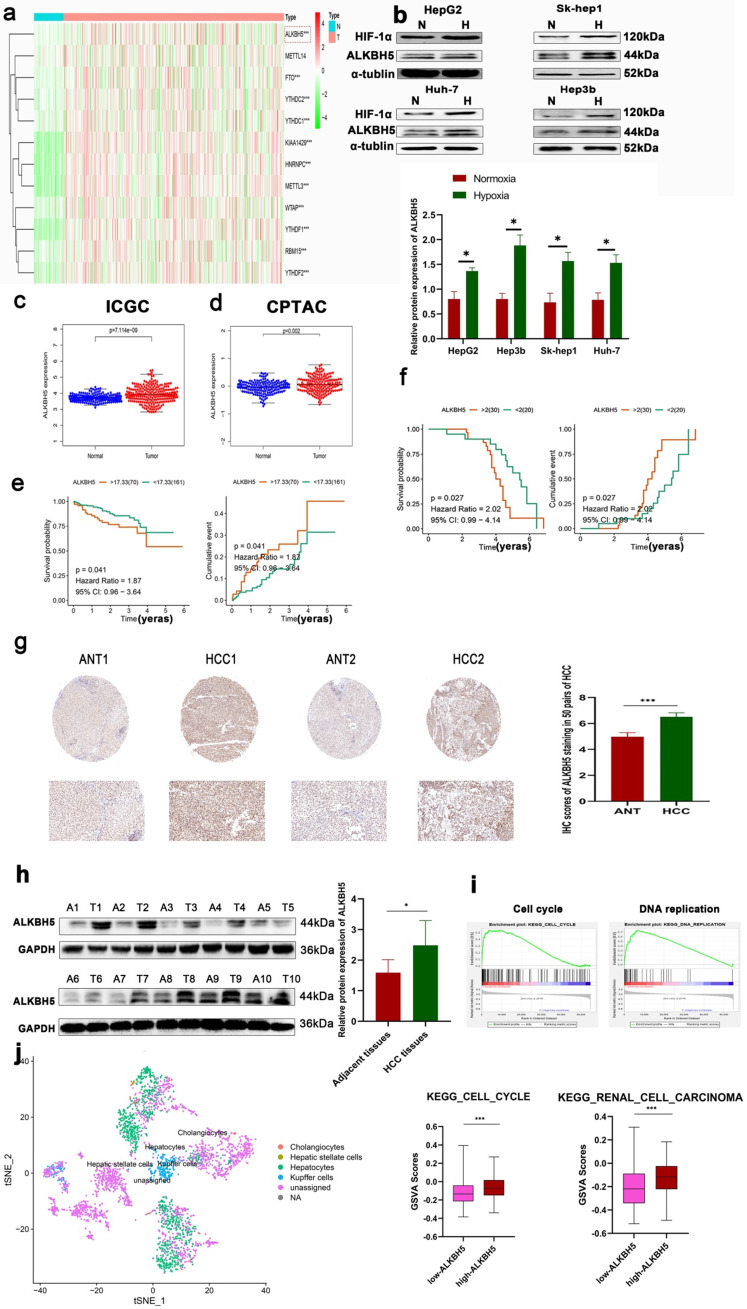
Figure 1.
ALKBH5 is upregulated in patients with HCC and closely related to poor prognosis. (a) Analysis of differentially expressed m6A-related genes in HCC based on the TCGA database. (b) Western blot analysis in HepG2, HUH-7, Sk-hep1 and Hep3b cells with or without hypoxia, n=3. (c) Analysis of differentially expressed ALKBH5 in HCC based on the ICGC database. (d) Analysis of differentially expressed ALKBH5 in HCC based on the CPTAC database. (e) Survival analysis of ALKBH5 in HCC based on the ICGC database. (f) Survival analysis of ALKBH5 in HCC based on the 50 pairs IHC tissues. (g) IHC assay for ALKBH5 in HCC tissue and adjacent normal tissue n=50. (h) Western blot analysis of ALKBH5 in HCC tissue and paracancerous tissue n=10. (i) GSEA of ALKBH5 in HCC based on TCGA-LIHC dataset the TCGA database. (j) GSVA analysis of ALKBH5 in independent liver tumor cells (Total hepatocytes were regards as HCC cells) based on the single-cell sequencing dataset GSE149614. *P < 0.05; **P < 0.01; ***P < 0.001. Comparisons between the HCC and adjacent tissues were analyzed by paired t test, and comparisons between the other two groups were analyzed by nonpaired t test. Comparisons among multiple groups were analyzed by one-way ANOVA. Cell experiments were repeated in triplicate. All data are presented as the means ± SEM.