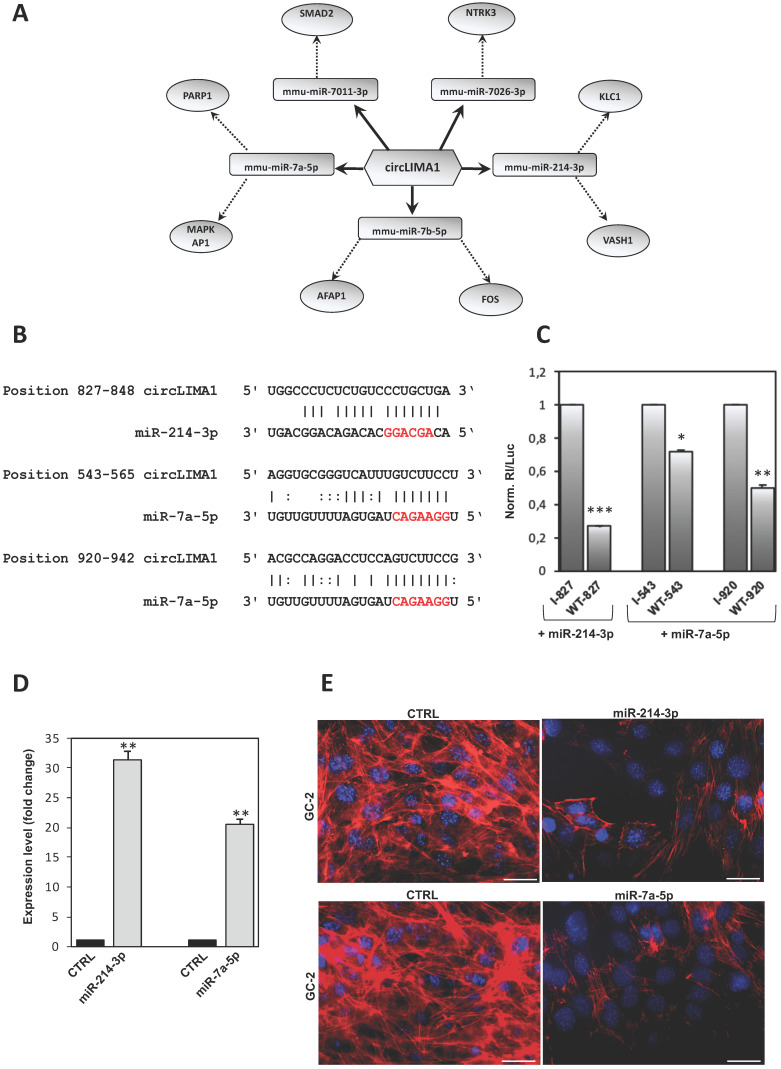
Figure 4.
CircLIMA1-dependent ceRNET validation and its involvement in the regulation of cytoskeletal actin. (A) Analysis of circLIMA1, up-regulated in CB1-/- compared to WT derived SPZ that tethers a group of miRNAs as targets, all involved in cytoskeleton-dependent pathways. Networks were built using Cytoscape. Hexagonal and rectangular symbols represent circRNAs and miRNAs, respectively. The arrow indicates the tethering activity of circRNAs toward miRNAs, while the dotted arrow indicates the pathways upstream of the miRNAs. (B) Prediction of miR-214-3p and miR-7a-5p binding site on circLIMA1 sequence; miRNA seed sequences are marked in red. (C) GC-2 cells were transfected with the luciferase-based reporter constructs containing the wild-type target sequence of miRNA (WT-starting nucleotide position number) or the control plasmids with inverted target sequence (I- starting nucleotide position number), along with 50 nM of the indicated miRNA mimic. After 48 hours, luciferase activities were recorded; the Renilla luciferase activity (Rl) was normalized to the firefly luciferase activity (Luc), whose coding sequence is carried by the same vectors; the values are reported as fold mean ± S.E.M. relative to RL/Luc recorded for transfection of controls (specific I construct + miRNA mimic) which were set to 1. Data are the mean ± S.E.M of three independent experiments, each with three data sets. *:p<0.05, **:p<0.01, ***:p<0.001. (D) qRT-PCR expression analysis of miR-214-3p and miR-7a-5p in GC-2 cells after 48 hours of miRNA mimic transfection. All qRT-PCR data are normalized using U6, expressed as fold change and reported as mean value ± S.E.M. ***: p<0.001. (E) Immunofluorescence analysis of F-actin by phalloidin staining (red) in GC-2 cells transfected with 50 nM of the indicated miRNA mimic. (n=3 different samples for each experimental group). Nuclei were labeled with DAPI (blue). Scale bar: 50 µm.