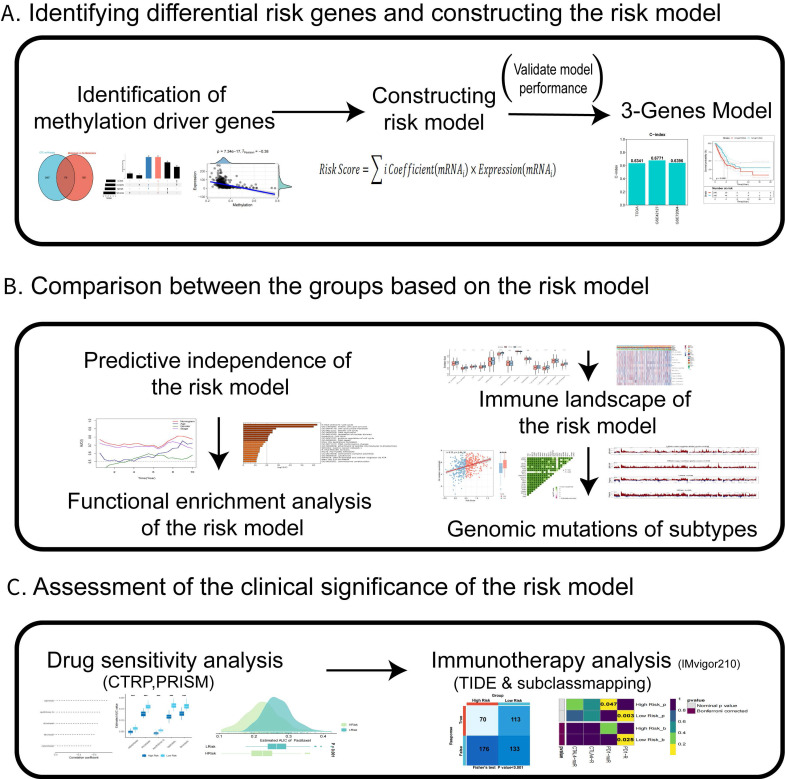
Figure 1.
The design and process of our study are shown in the flow chart. In the present study, We first identified differentially expressed genes in CTC and in situ tumors based on dataset GSE74639 and TCGA-LUAD dataset in metastatic patients versus non-metastatic patients, and obtained methylation probe information of differential genes to identify differentially methylated driver genes. Then, A risk model was constructed which comprised of three DMGs in CTCs of LUAD. Next, we systematically assessed the prognostic model's stability and accuracy in both the external validation and the training cohorts. The biological functions, TME, and genomic variations in the prognostic model were evaluated in detail. At last, the value of the prognostic model was determined and its clinical applicability in chemotherapy and immunotherapy of LUAD was evaluated.