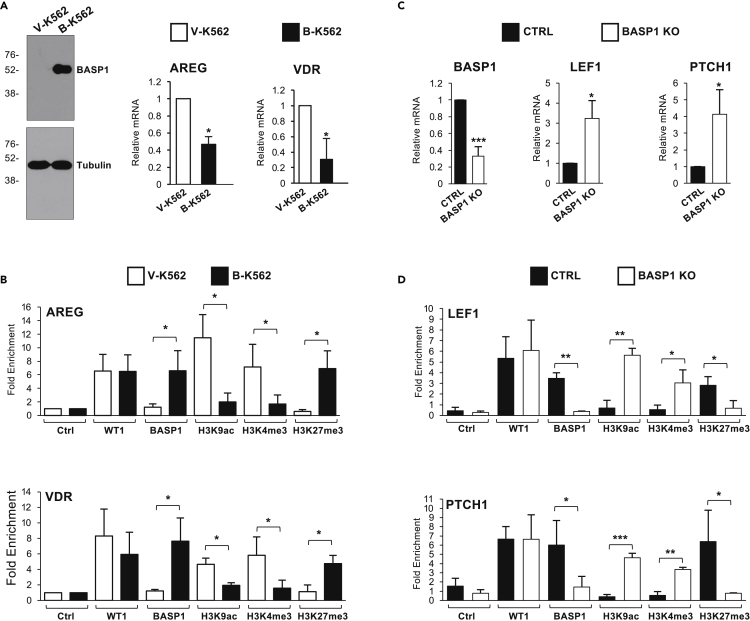
Figure 1.
BASP1 directs the removal of active histone modifications H3K9ac and H3K4me3 and the placement of repressive H3K27 trimethylation
(A) Immunoblotting of extracts prepared from V-K562 and B-K562 cells to confirm BASP1 expression. β-tubulin immunoblotting was performed as a loading control. Molecular weight markers are shown at left (kDa). cDNA was prepared from V-K562 cells and B-K562 cells and expression of AREG and VDR was quantitated relative to GAPDH. Data are SD from the mean (SDM) for three independent experiments. ∗ = p < 0.05 by students t-test.
(B) V-K562 or B-K562 cells were subjected to chromatin immunoprecipitation (ChIP) with control (ctrl) antibodies or antibodies against WT1, BASP1 or the histone modifications indicated. Data are presented as fold-enrichment over a control genomic region and error bars are SDM of three independent experiments. ∗ = p < 0.05 by students t-test.
(C) Krt8-BASP1-CRE mice were treated with tamoxifen for 8 days and then 7 days later taste buds were isolated and RNA prepared. cDNA was then used to monitor the expression of BASP1, LEF1, and PTCH1 compared to GAPDH. Error bars are SDM for three independent experiments. ∗ = p < 0.05 by students t-test.
(D) Mice were treated as in part C but isolated taste cells were subjected to chromatin immunoprecipitation with the antibodies indicated. Error bars are SDM for three independent experiments. ∗ = p < 0.05, ∗∗ = p < 0.01 and ∗∗∗ = p < 0.005 by students t-test.