Figure 4.
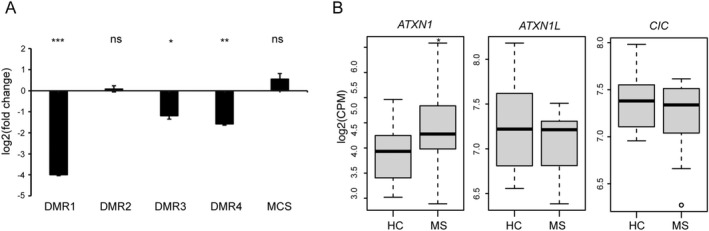
DNA methylation modulates ATXN1 expression. (A) Luciferase reporter vectors bearing the four DMRs were methylated in vitro and transfected in HeLa cells. The luciferase activity for each construct was tested 24 h after transfection and compared to the non‐methylated counterpart. A luciferase vector lacking the promoter region (MCS) was used as additional control. The data were expressed as log2 transformed fold change (log2FC) and plotted as means ± SE. Differences between experimental conditions were assessed by two‐tailed Student's t test. (B) The boxplots represent the RNA levels for ATXN1, ATXN1L, and CIC gene in B cells from MS patients (N = 23) and healthy controls (N = 14). The data were expressed as log2 transformed counts per million (log2CPM) and derived from RNA‐seq. Differential expression analysis was performed using the exact test implemented in the edgeR package. *p < 0.05, **p < 0.01, ***p < 0.001.
