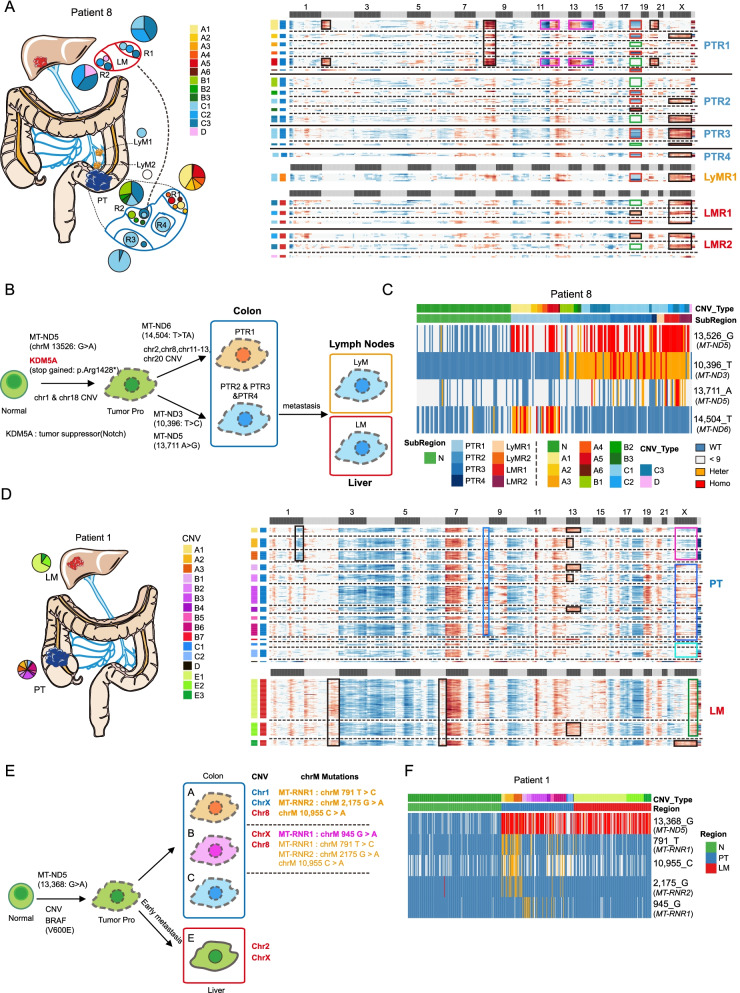
Fig. 5.
Possible presence of tumor precursor cells. A CNV pattern and sampling strategy of patient #8. N: adjacent normal tissues. PT: primary tumors and R1-R4 represent different regions of primary tumor. LyM: lymph node metastasis LyM1 and LyM2 represent two separated lymph node metastatic tumors. LM: liver metastasis. R1-R2 represents different regions of the liver metastatic tumors. The pie charts reflect the proportion of different subclones in each region. Copy number gain and copy number loss were indicated with red and blue respectively. Different tissues are divided by bold black solid lines, and different clones within the same tissue block are divided by black dashed lines. The squares above the heatmap represent different chromosomes, black squares represent odd-numbered chromosomes and chromosome X, and light gray squares represent even-numbered chromosomes and chromosome Y. The squares on the left of the heatmap represent different subclones, and the same color represents the same clone. The most obvious copy number differences between different clones were highlighted by different colored boxes. B The diagram showing the tumor metastasis path of patient #8. CNVs, somatic mutations and mitochondrial mutations are also shown in the diagram. The color of box represents different areas; blue, orange, and red box represent the primary tumor, lymphatic, and liver metastasis tumor respectively. Most of primary tumor cells have homozygous mutation at MT-ND5:13,536G>A mutation; thus, we speculated that there may be a group of tumor progenitor cells that we did not capture, which have MT-ND5(13,536G>A) mutation. With the development of the tumor, this group of tumor progenitor cells have additional mutations at MT-ND6(14,504 T>TA) and MT-ND3(10,396T>C) respectively, thus forming two clones (PTR1 A clones and PTR2-PTR4 B&C clones). Then the clones with MT-ND3(10,396T>C) mutation were further metastasized to the liver. C Heatmap showing selected mitochondrial mutations of patient #8. Almost all tumor cells have a chrM:13,526 homozygous mutation. Cells from the PTR1 (primary tumor region 1) have a chrM:14,504 mutation, and cells from PTR2-PTR4 and lymph node metastasis have a chrM:10,396 mutation. Blue represents wild-type and gray represents read depth lower than 9. Orange represent heterozygous mutation and red represent homozygous mutation. The bars above the heatmap shows the CNV subclones and tissue origin of the cells. D CNV pattern and sampling strategy of patient #1. PT: primary tumors. LM: liver metastasis. Copy number gain and copy number loss were indicated with red and blue respectively. Different tissues were divided by bold black solid lines, and different clones within the same tissue block were divided by black dashed lines. The squares above the heatmap represent different chromosomes, black squares represent odd-numbered chromosomes and chromosome X, and light gray squares represent even-numbered chromosomes and chromosome Y. The squares on the left of the heatmap represent different subclones, and the same color represents the same clone. The most obvious copy number differences between different clones are highlighted by different colored boxes. The black square in chromosome 1 shows A1–A3 subclone-specific CNV. The black squares in chromosome 2 and chromosome 6 show additional CNVs of LM (E1–E3 clone-specific clones) compared with PT. E The diagram showing the tumor metastasis path of patient #1. CNVs, somatic mutations and mitochondrial mutations are also shown in the diagram. F Heatmap showing selected mitochondrial mutations of patient #1. All tumor cells have homozygous mutation at 13,368 point site. N: adjacent normal tissue. PT: primary tumor. LM: liver metastasis