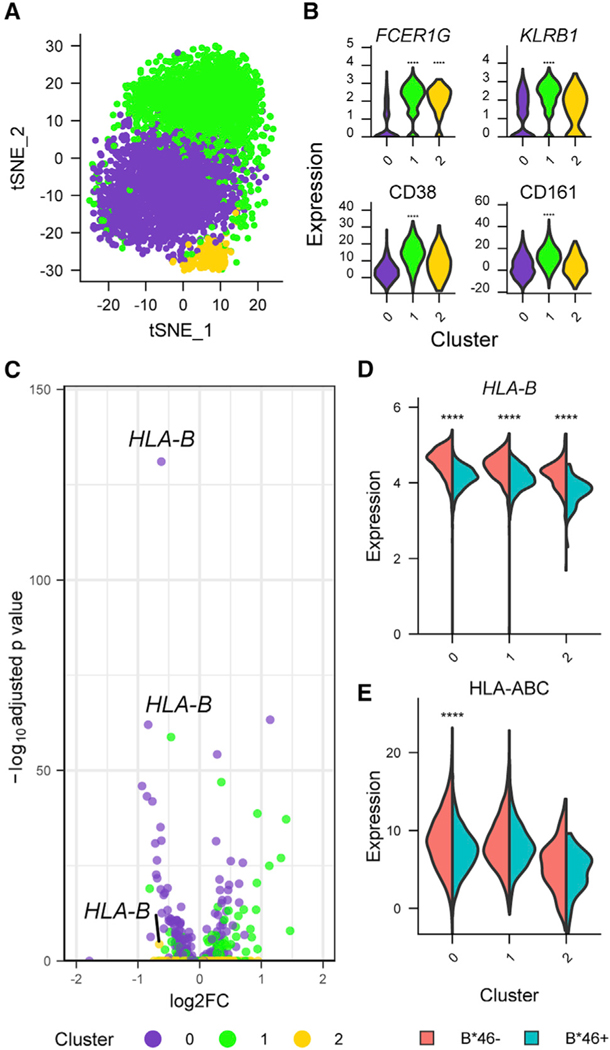
Figure 5. Single-cell CITE-seq data reveal that the same NK population that exhibited a distinct transcriptional phenotype associated with B*46 after infection is also present in the absence of HIV infection.
(A) CITE-seq data from PWOH (n = 9) identified three NK cell clusters.
(B) Gene and protein markers of the specific NK cell cluster differentiated by B*46 during HIV infection in the current three NK cell populations. Statistical significance is denoted for comparisons between one cluster with the combined two other clusters. ****p < 0.0001.
(C) HLA-B is the most differentially expressed gene comparing B*46+ versus B*46− groups.
(D and E) (D) Gene (HLA-B) and (E) protein (HLA class I expression using W6/32 antibody) expression in all clusters were higher in the B*46− group. Statistical significance is denoted for comparisons between the groups within a specific cluster. Statistical significance for gene and protein clusters were determined by the Wilcoxon rank-sum and likelihood ratio tests, respectively. ****p < 0.0001.