Figure 6.
Mpe1 is globally required for timely transcription termination
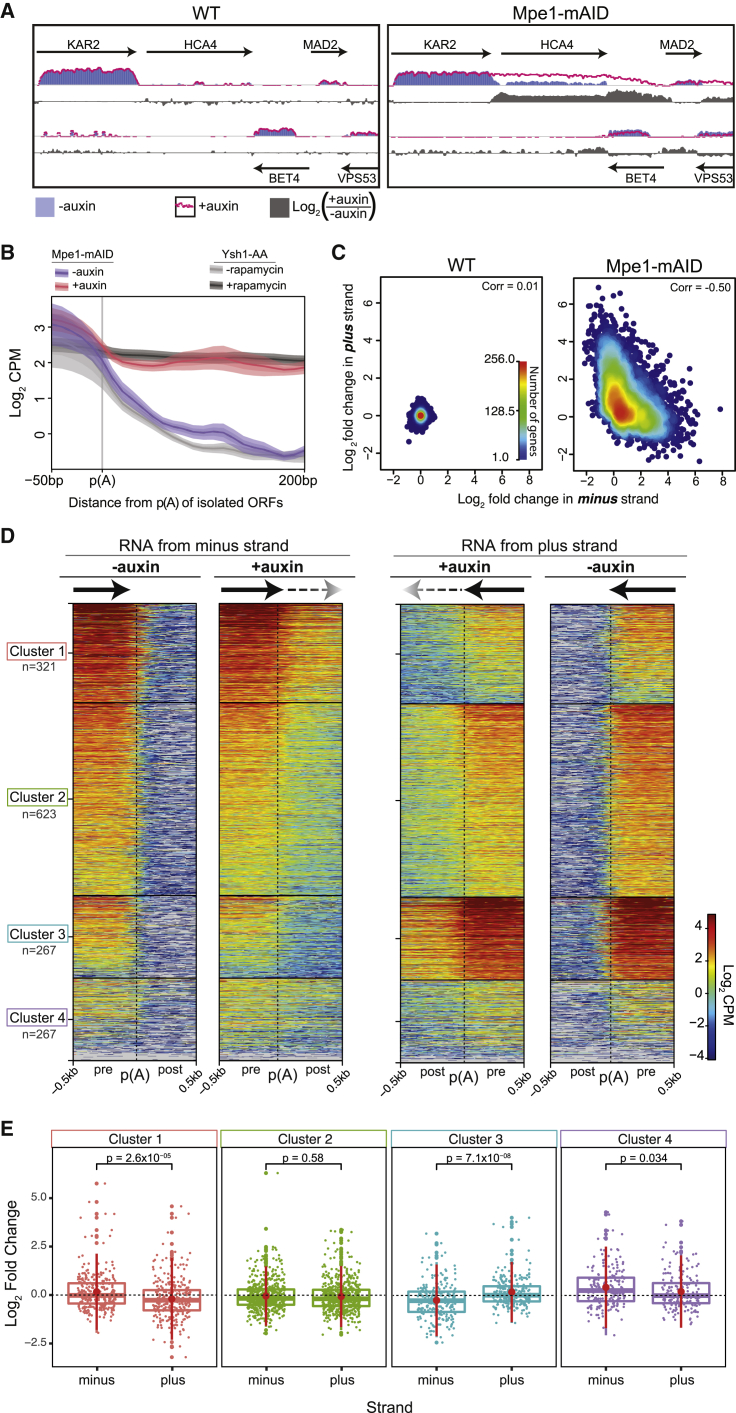
(A) Representative genomic snapshots of strand-specific nascent RNA-seq from WT (left) or Mpe1-mAID (right) yeast, either untreated (blue bars) or treated with auxin (magenta trace). The log2-fold change in nascent RNA upon the addition of auxin is in gray. Arrows represent protein-coding genes.
(B) Metagene plots of nascent RNA at the polyadenylation site (poly(A)) from the Mpe1-mAID cells treated with auxin (magenta) or untreated (blue). Nascent RNA from Ysh1 anchor away cells (Ysh1-AA), where Ysh1 was depleted (+rapamycin, dark gray) or not depleted from the nucleus (−rapamycin, light gray) is also shown. Ysh1 depletion data were obtained in a previous study (Baejen et al., 2017) and re-analyzed here. Selected genes are ≥200 bp from neighboring ORFs (n = 931 genes). Center line of each curve represents average signal; shaded area is 95% confidence interval.
(C) Density scatter plot of changes in nascent RNA synthesis in WT or Mpe1-mAID cells upon the addition of auxin. Values correspond to strand-specific log2-fold change per gene and corresponding position on the opposite strand.
(D) k-means clustering of strand-specific nascent RNA before and after auxin treatment in Mpe1-mAID cells. Data are shown for a 1-kb window centered around the polyadenylation site (poly(A)) of 1,478 convergent gene pairs. Transcription directionality is indicated with arrows. The number of genes in each cluster (n) is indicated. CPM, counts per million.
(E) Log2-fold change in nascent RNA upon Mpe1 depletion for the genes in each of the clusters in (D) on the minus and plus strands. Dots represent the log2-fold change for each gene. Large dots represent outliers within each distribution. p values are from pairwise Student’s t test. Middle horizontal line in each boxplot represents the median, and the boxes show the interquartile range. Number of genes (n) for each cluster is shown in (D).
See also Figures S6 and S7.