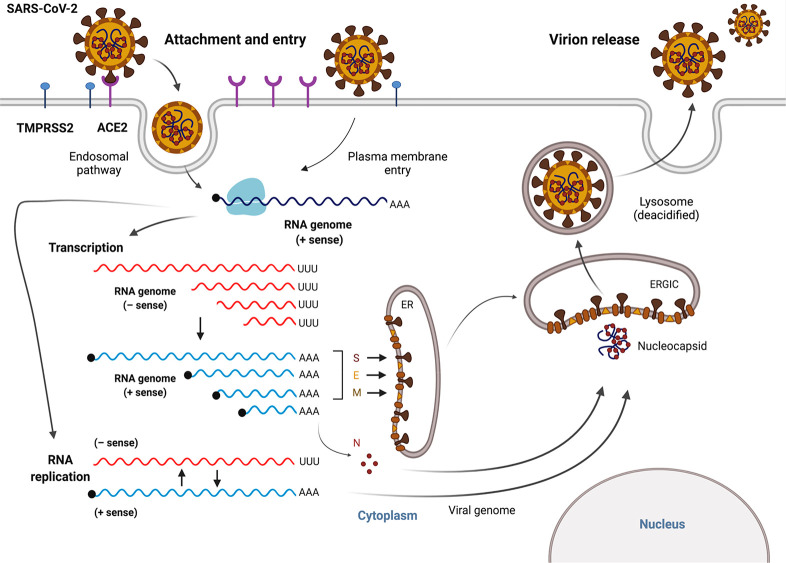
Figure 3.
The SARS-CoV-2 replication cycle. SARS-CoV-2 enters the host cell via an endosomal pathway or through fusion of the viral envelope with the host cell membrane. Briefly, viral entry is initiated by binding of the RBD of the spike (S) protein to the human host cell receptor (ACE2). After the RBD–receptor interaction, the S protein undergoes proteolytic cleavage, which can be catalyzed by several host proteases, such as TMPRSS2, furin, and cathepsin B/L. Following viral entry, SARS-CoV-2 releases its genomic RNA into the cytoplasm and utilizes both the host’s and its own enzymatic machinery to replicate its genetic material and assemble new viral particles. The viral RNA genome is first translated into viral replicase polyproteins (pp1a and pp1ab), which are then cleaved into 16 nsps. In the process of genome replication and transcription mediated by the replication–transcription complex (RTC), the negative-sense (− sense) genomic RNA is synthesized and used as a template to generate a positive-sense (+ sense) genomic RNA and subgenomic RNAs. Viral assembly is aided by the interaction between viral genomic RNA and structural proteins located in the endoplasmic reticulum (ER) and ER–Golgi intermediate compartment (ERGIC). Finally, these virions are released to the plasma membrane via deacidified lysosomes and secreted from the infected cell via exocytosis. This figure was created with Biorender.com.