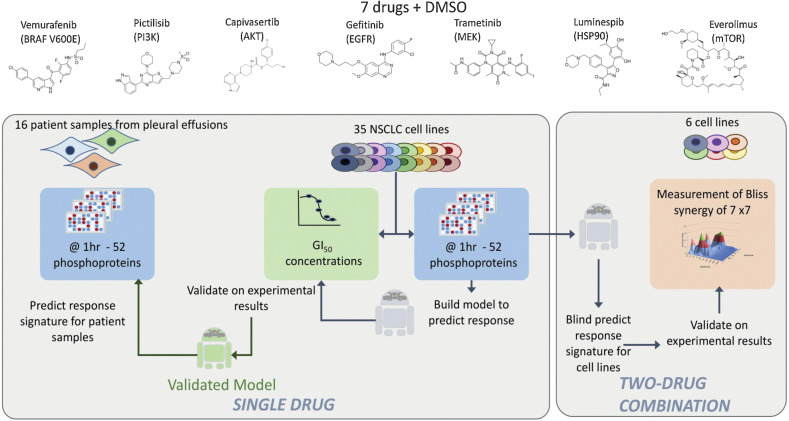
Figure 1.
Experimental design. Single-drug evaluation: A library of 7 targeted anticancer drugs was used. First, GI50 concentrations were determined in a panel of 35 NSCLC cell lines with diverse genetic backgrounds (44). Second, phosphoproteomic changes of 52 selected proteins were measured after 1 hour of drug exposure of the drugs at clinically relevant concentrations adjusted for protein binding and DMSO controls were measured. The phosphoproteomic protein changes were used to train machine learning predictors of sensitivity, and validated using 100-fold cross validation with a rotating set of 15% leave out for validation and 85% for training (see Materials and Methods). The same phosphoproteomic measurements were also carried out in 16 patient samples obtained from pleural effusions producing profiles that can be fed into the predictive model to predict likely response to each drug of the individual patient samples. Two-drug combination: A novel machine learning method (environmental perturbation score) using dynamic phosphoprotein data 35 cell lines exposed to the 7 drugs was used to predict combinations. All pair wise two-drug combinations (7 individual drugs) were tested in 6 representative NSCLC cell lines and Bliss synergy was calculated for all combinations. The predicted results from the environmental perturbation score was compared with the experimentally validated results.