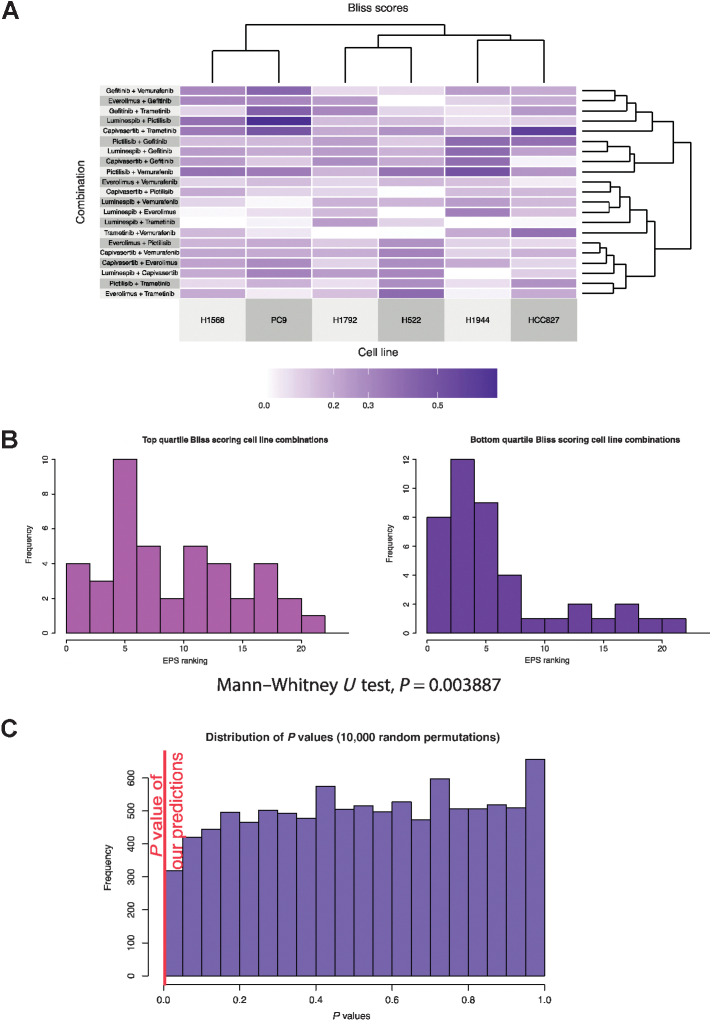
Figure 5.
Experimental and predicted combinations. A, Clustered heatmap of Bliss synergy scores was experimentally measured for six cell lines treated with 21 two-drug combinations. B, Histogram representing the EPS rankings of nodes of targets of drugs in the top 25% highest Bliss synergy scores, that is, “most synergistic” (left), or the EPS rankings of nodes of targets of drugs in the 25% lowest Bliss synergy scores, that is, “least synergistic” (right). There is a significant bias toward higher EPS rankings for the most synergistic drug targets, with a significant Mann–Whitney U test P value of 0.0038875, indicating a biased distribution of rankings. C, Simulation of the Mann–Whitney U test P values obtained from 10,000-fold random permutations of EPS ranking, demonstrating the robustness of this P value.