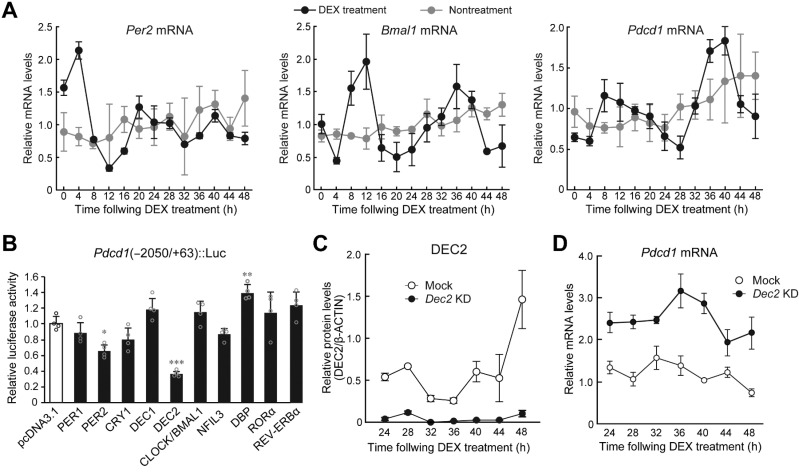
Figure 2.
DEC2 regulates the circadian expression of Pdcd1 mRNA in macrophages. A, Temporal mRNA expression profiles of Per2, Bmal1, and Pdcd1 in RAW264.7 cells, whose circadian clocks were synchronized by treatment with 100 nmol/L DEX for 2 hours. Nontreatment cells were set as a nonsynchronized control. Data were normalized by the levels of 18S rRNA and the mean of each group was set at 1.0. Values are the mean with SD (n = 3). There were significant time-dependent variations in Per2, Bmal1, and Pdcd1 in DEX treatment group (F12, 26 = 53.225, P < 0.001 for Per2; F12, 26 = 12.609, P < 0.001 for Bmal1; F12, 26 = 18.874, P < 0.001 for Pdcd1; one-way ANOVA). B, DEC2 negatively regulates the transcription of the Pdcd1 gene. RAW264.7 cells were cotransfected with Pdcd1(-2050/+63)::Luc, and expression vectors for PER1, PER2, CRY1, DEC1, DEC2, CLOCK/BMAL1, NFIL3, DBP, RORα, and REV-ERBα. The values are the mean with SD (n = 3). The value of empty vector (pcDNA3.1)-transfected RAW264.7 cells was set at 1.0. *, P < 0.05; **, P < 0.01; ***, P < 0.001; significant difference from empty vector (pcDNA3.1)-transfected groups (F10,33 = 18.109, P < 0.001; ANOVA with the Tukey Kramer post hoc test). C, Temporal expression profiles of DEC2 protein in mock-transduced and Dec2 KD RAW264.7 cells, whose circadian clocks were synchronized by treatment with 100 nmol/L DEX. Data were normalized by the β-ACTIN levels. Values are the mean with SD (n = 3). (F1, 28 = 216.110, P < 0.01 for group; F6,28 = 17.930, P < 0.01 for time point; F6,28 = 12.966, P < 0.01 for time point×group; two-way ANOVA). D, Temporal expression profiles of Pdcd1 mRNA in mock-transduced and Dec2 KD RAW264.7 cells, whose circadian clocks were synchronized by treatment with 100 nmol/L DEX. Data were normalized by the levels of 18S rRNA. Values are the mean with SD (n = 3). (F1,28 = 323.663, P < 0.01 for group; F6,28 = 8.459, P = 0.016 for time point; F6,28 = 4.890, P < 0.01 for time point×group; two-way ANOVA).