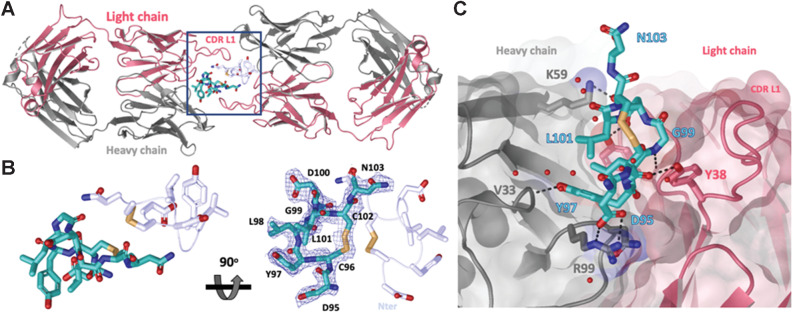
Figure 3.
Crystal structure of LY6G6D–1G4 Fab complex at 2.2 Å resolution suggests Fab binds an epitope located close to the membrane. A, Ribbon diagram of one crystallographic dimer shown from a top view. The complex crystallized in space group P 1 21 1 with two dimers per asymmetric unit; the four complexes within the asymmetric unit superimpose with a root–mean–square deviation of less than 0.5 Å. The light and heavy chains from the Fab are colored in red and gray, respectively. The LY6G6D peptides (Asp95-Asn103) are shown in cyan and light blue and side chains are shown as sticks. Disulfides are shown in yellow (PDB accession pending). B, Close-up view of LY6G6D peptides within dimer (from highlighted box in A). mFo−DFc omit map for one LY6G6D peptide is shown in blue mesh contoured at 1σ. Both main and side chain atoms are shown; side chains density for most of the residues is clearly defined. For clarity, only the side chains of the symmetry-related peptide are shown in stick. Two views rotated by 90 degrees are depicted. C, Details of molecular interactions between the LY6G6D peptide (shown in sticks and colored in cyan) and its proximal Fab (surface representation, key CDR side chains involved in binding are shown in sticks). The peptide binds within a cleft between the light and heavy CDRs of the Fab. Water is represented as red spheres and contacts (hydrogen bonds and salt bridges) as black dotted lines.