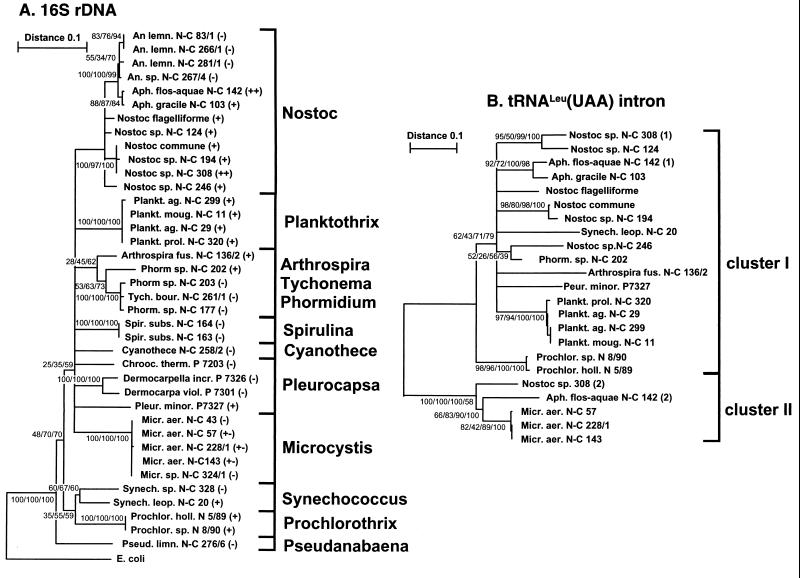
FIG. 6.
Evolutionary trees for 16S rDNA (A) and tRNAUAALeu introns sequences (B). The trees were built using the neighbor-joining, maximum-parsimony, and maximum-likelihood methods. The trees shown in panels A and B (based on 489 and 346 aligned positions, respectively) are consensus trees for the branches supported by ≥25% of the bootstrap trees in all of the phylogenetic methods tested. The genetic distances between two strains are Kimura distances expressed in substitutions per nucleotide in the neighbor-joining tree. Numbers at the nodes (minimal evolution [for the intron tree]/maximum likelihood/maximum parsimony/neighbor joining) indicate the percentage of the bootstrap trees in which the cluster descending from the node was found. In panel A, the presence and absence of introns are indicated with (+) and (−), respectively. The respective clone numbers (N-C, NIVA-CYA; N, NIVA; P, Pasteur) are given for each branch. Species abbreviations: An. lemn., Anabaena lemmermannii; Tych. bour., Tychonema bourrellyi; Spir. subs., Spirulina subsalsa; Cyanothece, Cyanothece aeruginosa; Chrooc. therm., Chroococcidiopsis thermalis; Dermocarpella incr., Dermocarpella incrassata; Dermocarpella viol., Dermocarpella violacea; Pseud. limn., Pseudanabaena limnetica. All other species are abbreviated as in Fig. 2 and 3.