Figure 1.
RET::GRB2 fusion is detected as a somatic event in a pheochromocytoma
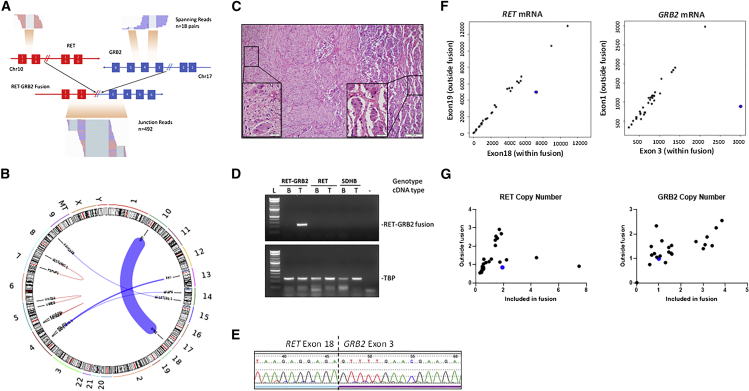
(A) Representation of the region spanning the RET exon 18 (left panel) and GRB2 exon 3 (right panel) in a pheochromocytoma predicted to carry the RET::GRB2 fusion, indicating the number of spanning reads (n = 18) and junction reads (n = 492) detected by RNA-seq. Chromosome (chr) location is indicated for each gene.
(B) Chromosomes (chrs) plotted as ideograms around the outside of the circle (CIRCOS) plot depicting the putative recombination between RET (chr10) and GRB2 (chr 17). The thickness of the connecting line represents the depth of read coverage.
(C) Hematoxylin and eosin (H&E) staining of the primary composite tumor comprised of a predominant pattern of pheochromocytoma (right) combined with a focal area of ganglioneuroma (left); magnification 100×, scale bar, 100 μm (inset, 400×, scale bar, 20 μm).
(D) Agarose gel of PCR products spanning the RET::GRB2 fusion transcript in tumor but not matched blood (leukocyte) cDNA; two other paired blood/tumor cDNA samples from patients with a RET mutation and a SDHB germline mutation, respectively, show no product (top); housekeeping gene TBP is shown (bottom). These results were repeated three times.
(E) Sanger sequencing of the product shown in (D), displaying RET sequence (exon 18) merged with GRB2 (exon 3). This experiment was repeated three times.
(F) RET (left) and GRB2 (right) mRNA expression obtained from high-throughput sequencing (HT-seq) counts of RNA-seq from 30 pheochromocytomas/paragangliomas (PPGLs; dots), depicting an exon within the fusion (x axis) and one outside the fusion (y axis). RET::GRB2-positive tumor is indicated in blue.
(G) DNA-based quantitative real-time PCR assay for copy-number assessment of the RET and GRB2 genes using primers targeting the area within (x axis) or outside the fusion (y axis) of a set of 14 PPGLs of various genotypes (dots). This experiment was repeated three times in duplicate samples.