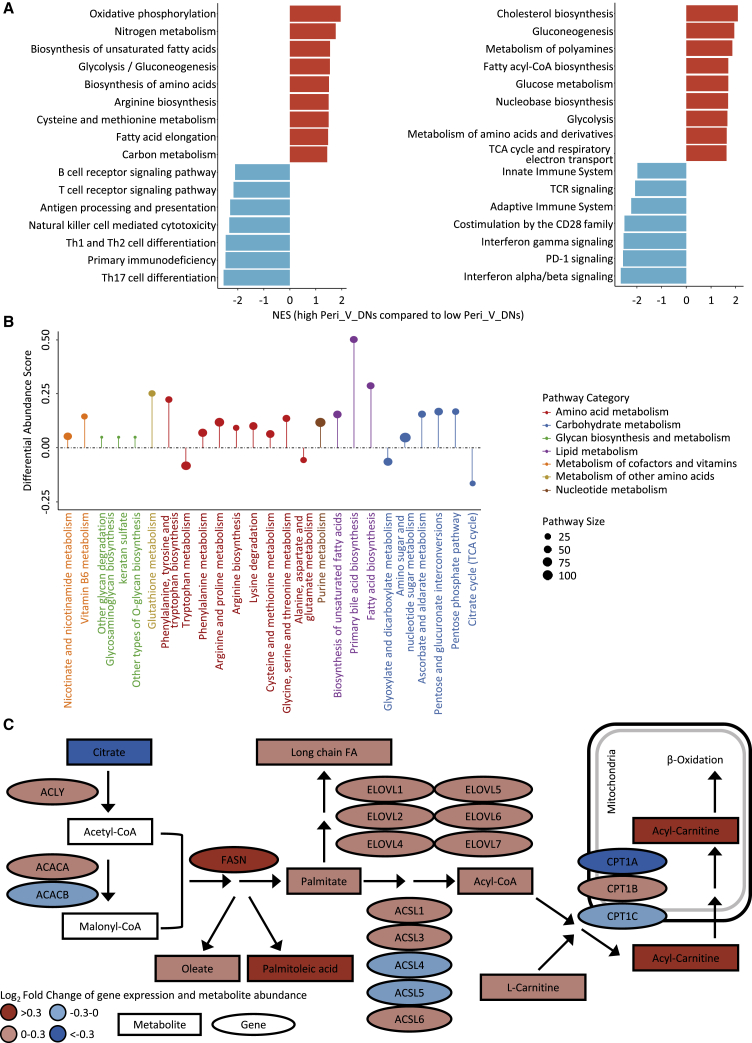
Figure 4.
Identification of differentially expressed pathways and transcriptomic-metabolomic integrative analysis
(A) Enrichment of pathways in high Peri_V_DN group compared with low Peri_V_DN group using GSEA (left panel based on KEGG; right panel based on Reactome).
(B) A pathway-based analysis of metabolomic changes between Peri_V_DN groups. The differential abundance (DA) score captured the overall change in a metabolic pathway. A score of 1 indicated that all metabolites in this pathway increased in high Peri_V_DN group compared with low Peri_V_DN group, and a score of −1 indicated that all metabolites in this pathway decreased.
(C) Transcriptomics and metabolomics distinctions in fatty acid biosynthesis pathway between Peri_V_DN groups. Log2-fold changes of mRNA expression levels and metabolite abundances in high Peri_V_DN tumor samples compared with low Peri_V_DN tumor samples were demonstrated.
CoA, coenzyme A; FA, fatty acid; GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score; TCA, tricarboxylic acid; TCR, T cell receptor. See also Figure S4 and Tables S5, S6, S7, and S8.