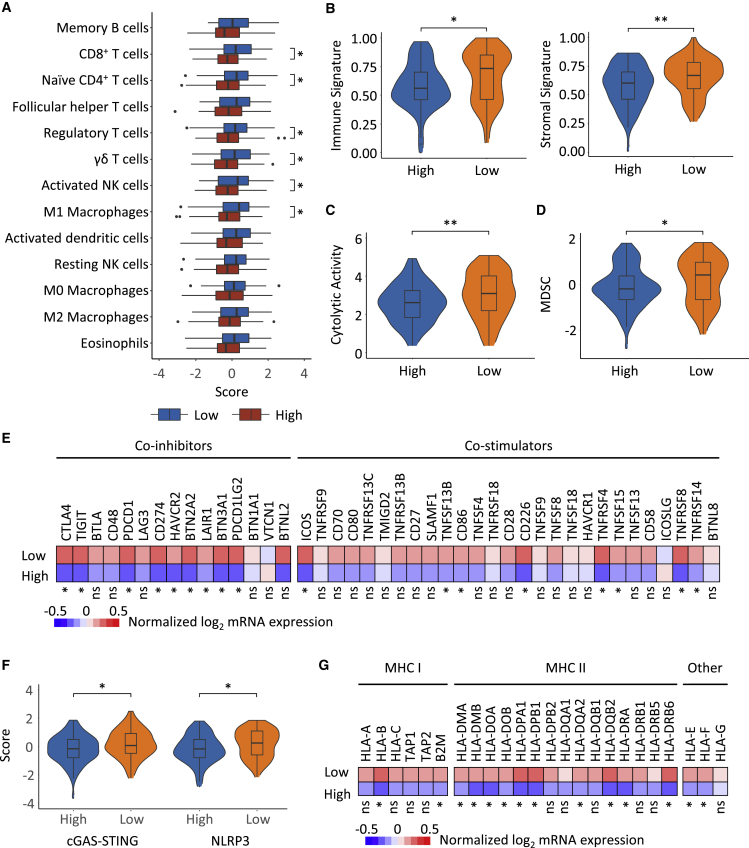
Figure 5.
Landscape of the tumor microenvironment of the Peri_V_DN groups and distinct escape mechanisms
(A) Differences in the abundance of immune cell types in high Peri_V_DN group compared with low Peri_V_DN group.
(B) Scores of the immune signature (left) and stromal signature (right) inferred by ESTIMATE22 between Peri_V_DN groups.
(C) Comparison of cytolytic activity showed higher effector immune cell activity between Peri_V_DN groups.
(D) Comparison of the abundance of MDSCs between Peri_V_DN groups.
(E) Normalized mRNA expression levels of immune co-inhibitors and co-stimulators between Peri_V_DN groups.
(F) Signature scores of two innate immunity-sensing pathways, cGAS-STING and the NLRP3 inflammasome, between Peri_V_DN groups.
(G) Normalized mRNA expression levels of MHC molecules between Peri_V_DN groups. In total, 167 samples with transcriptomic data were included for analysis.
∗∗0.001 < p ≤ 0.01; ∗0.01 < p ≤ 0.05; ns, p > 0.05. GSVA, gene set variation analysis; MDSC, myeloid-derived suppressor cell; ssGSEA, single-sample gene set enrichment analysis.