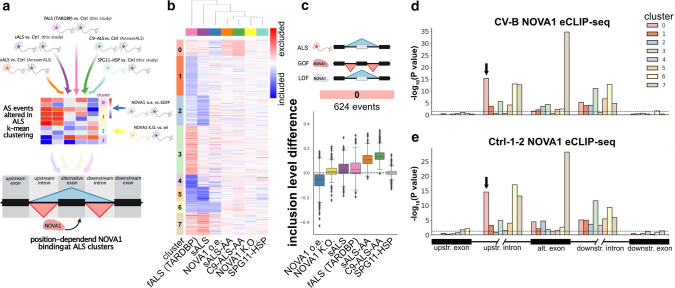
Fig. 5.
Aberrant NOVA1 states in ALS exhibit gain- and loss-of-function features a Schematic illustrating analysis strategy to identify patterns of altered NOVA1 function in ALS iPSC-derived MNs. b Heatmap of all significant AS events observed in the four ALS datasets. Inclusion level difference for all depicted datasets were clustered using k-means clustering into 8 distinct clusters. Blue denotes inclusion (max: −0.6), red denotes exclusion (max: 0.6). Datasets are clustered using Euclidean distance metric. c Boxplot of inclusion level differences in clusters 0 (sALS vs. Ctrl: purple; fALS vs. Ctrl: pink; sALS vs. Ctrl (AA): orange; C9-ALS vs. Ctrl (AA): green; SPG11-HSP vs. Ctrl: cyan; NOVA1 vs. EGFP overexpression: blue; NOVA1 wt vs. NOVA1 K.O.: yellow). Ideogram on top illustrates the direction of the most prominent observed change in ALS, NOVA1 gain of function (GOF) and NOVA1 loss of function (LOF) (from top to bottom). d Bar blot illustrating significance (negative log10 scale) of CV-B NOVA1 eCLIP-seq binding site enrichment at AS events in the 8 detected AS event clusters at upstream exon, upstream intron, alternative exon, downstream intron, and downstream exon. Statistical significance was calculated with hypergeometric test against all detected AS events in the four ALS datasets as a background. Dashed line represented P value = 0.05. Arrow depicts most significant enrichment in cluster 0. e Bar blot of significance (negative log10 scale) of Ctrl-1-2 NOVA1 eCLIP-seq binding site enrichment at AS events in the 8 detected AS event clusters at upstream exon, upstream intron, alternative exon, downstream intron and downstream exon. Statistical significance was calculated with hypergeometric test against all detected AS events in the four ALS datasets as a background. Dashed line represents P value = 0.05. Arrow depicts most significant enrichment in cluster 0