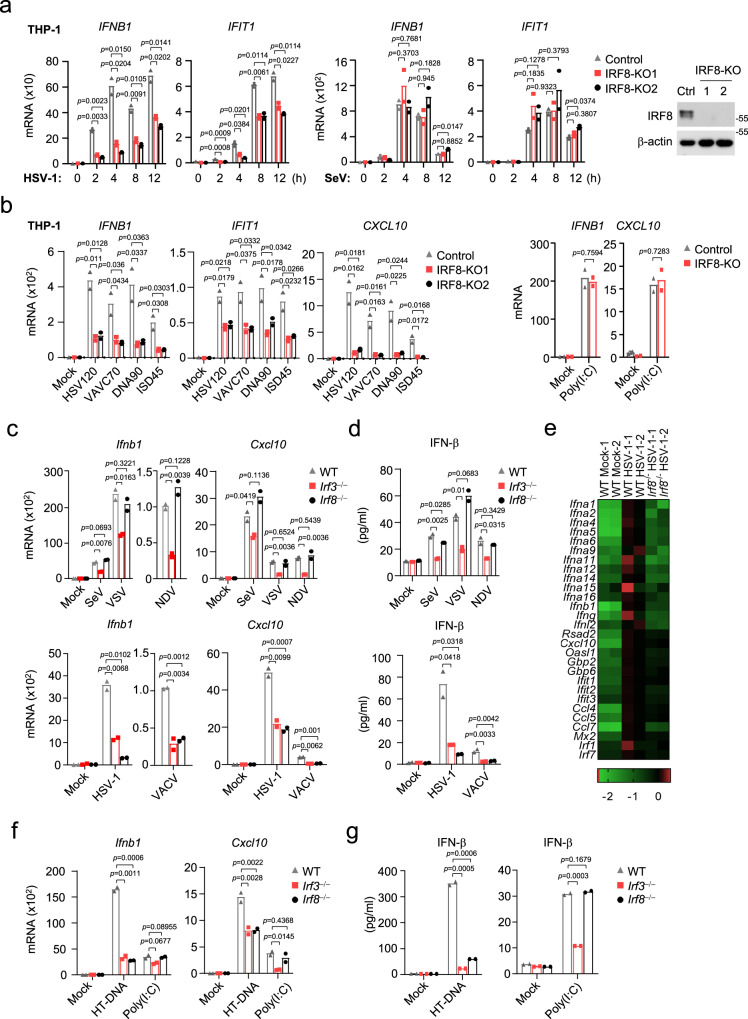
Fig. 1. IRF8-deficiency inhibits DNA-triggered signaling in monocytes.
a qPCR analysis of IFNB1 and IFIT1 mRNAs in IRF8 knockout (IRF8-KO-1 and IRF8-KO-2) or control THP-1 cells (transduced with the lenti-CRISPR-V2 containing a control gRNA) infected with SeV or HSV-1 for the indicated times. The knockout efficiencies of IRF8 were shown by immunoblots. b qPCR analysis of mRNA levels of the indicated genes in IRF8-KO or control THP-1 cells transfected with the indicated nucleic acids for 4 h. qPCR analysis of Ifnb1 and Cxcl10 mRNAs (c) and ELISA analysis of Ifn-β secretion (d) in WT, Irf8−/− and Irf3−/− BMDMs left uninfected or infected with the indicated viruses for 4 h. e Heat map of hierarchical clustering analysis of RNA-sequencing data from WT and Irf8−/− BMDMs left uninfected or infected with HSV-1 for 4 h. The scale (log10) represents the changes in gene expression among different samples. qPCR analysis of Ifnb1 and Cxcl10 mRNAs (f) and ELISA analysis of Ifn-β secretion (g) in WT, Irf8−/− and Irf3−/− BMDMs transfected with the indicated nucleic acids for 4 h. Data in a–d, f and g are presented as mean. n = 2 technical replicates. Data were analyzed by unpaired two-tailed Student’s t-test. Source data are provided as a Source data file.