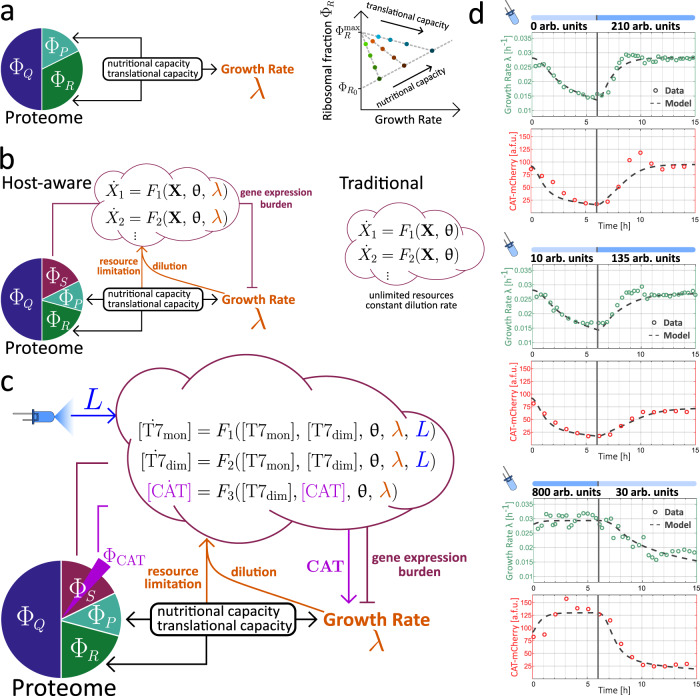
Fig. 4. Host-aware modeling framework applied to the photophilic strain.
a In proteome-partition models, empirical correlations between growth rate and ribosomal mass fraction arise from a balance between co-regulated sectors of the proteome: a fixed, house-keeping fraction (ΦQ) and two flexible sectors, the ribosomal fraction (ΦR) and the catabolic fraction (ΦP). The give-and-take regulation of the latter two sectors determines the cellular growth rate, mediating adaptation to environmental conditions characterized by nutrient quality and translational capacity. Figure adapted from refs. 47 and 54. b In our approach to host-aware modeling, the dynamics of arbitrary synthetic genetic circuits, described by a system of ODEs, are embedded into a proteome-partition framework that captures the physiological response of the cell. In contrast to a traditional ODE model, which does not consider the physiological adaptation of the host's growth rate, gene-expression burden caused by the circuit or limitations in the host's gene-expression resources, the host-aware framework seamlessly incorporates all of these host-circuit interactions without the need for extra free parameters. c Host-aware modeling framework applied to the photophilic strain. Blue-light intensity enters as an external parameter and the growth-modulating effect of expressing CAT in the presence of chloramphenicol introduces a direct interaction between the circuit and the host's growth rate. d Dynamic upshift and downshift experiments used to determine the best parameter values for the host-aware model of the photophilic strain. The parameterized model simultaneously recapitulates the dynamics of both cellular growth rate and resistance expression.
Source data are provided as a Source Data file.