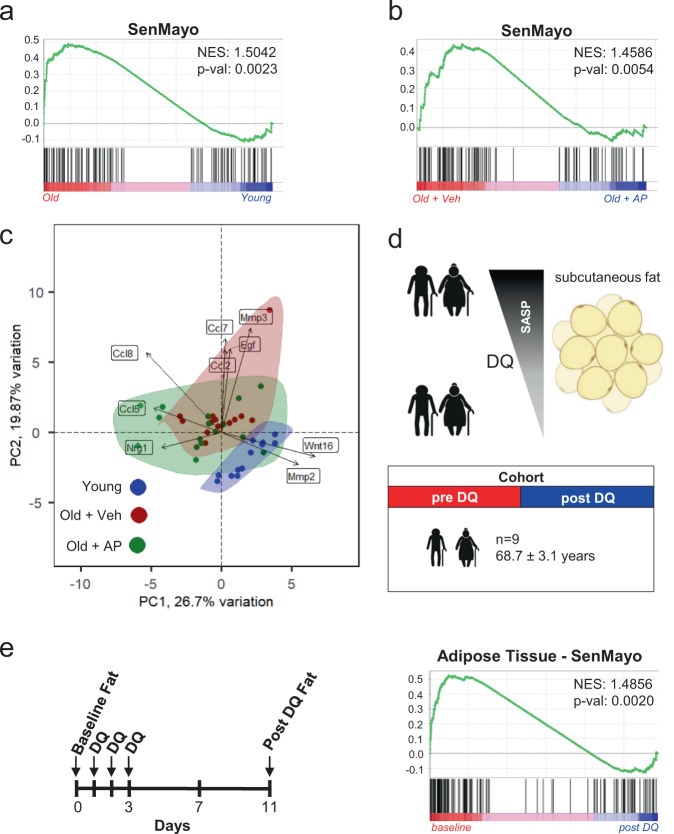
Fig. 3. The SenMayo gene set tracks genetic and pharmacologic clearance of senescent cells.
a The SenMayo panel successfully indicated aging in bone in mice (p value = 0.0023), n = 25 (12 young, 13 old (all ♀). Nominal p value, calculated as two-sided t-test, no adjustment since only one gene set was tested; b The elimination of p16Ink4a-expressing senescent cells by AP20187 administration was shown previously to reverse the aging bone phenotype37. The SenMayo gene set successfully demonstrated the significant reversal of the aging phenotype at the gene expression level upon the elimination of p16Ink4a-expressing senescent cells (p = 0.0054), n = 29 (13 Veh, 16 AP (all ♀). Nominal p value, calculated as two-sided t-test, no adjustment since only one gene set was tested; c By specifically using the expression patterns of the SenMayo gene set, our bone RNA-seq revealed no similarities in gene expression patterns between young (blue) and old + veh (red) treated mice, and a substantial overlap of expression profiles of old + AP (green) mice with young mice. The highlighted genes represent variables, and the arrows drawn from the origin indicate their “weight” in different directions, according to the theories of Gabriel103; d We used a previously published mRNA-seq dataset from human adipose tissue of our group24, 39, to evaluate changes in SenMayo following D + Q treatment. Adipose tissue was collected before and 11 days after three days of oral D + Q treatment. Figure was designed using Biorender.com; e Using SenMayo, there was a reduction of SenMayo (p = 0.002) in the subcutaneous fat samples in the subjects treated with D + Q, consistent with a reduction in senescent cell burden following D + Q treatment (n = 9 (7 ♂, 2 ♀)). Nominal p value, calculated as two-sided t-test, no adjustment since only one gene set was tested. Source data are provided as a Source Data file.