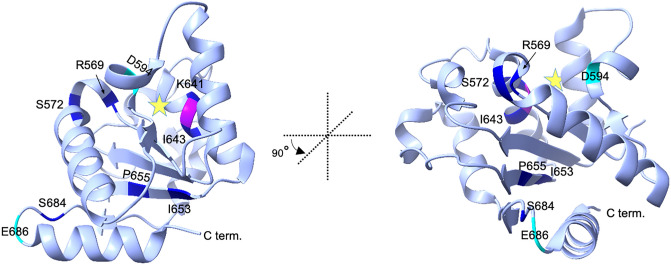
Figure 6.
Locations of missense variation that confer NADase LoF in the SARM1 TIR domain crystal structure. Ribbon diagram of the reported crystal structure of the human SARM1 TIR domain showing the locations of residues that are altered by the natural missense coding variants identified in this study to cause LoF. The structure presented is based on PDB ID:6O0R (bound to ribose) with the position of the predicted NAD substrate-binding pocket indicated by a star. Two views are shown differing by a rotation of 90° anti-clockwise along the vertical axis. The locations of residues that are altered to confer basic LoF are shown in dark blue and those that are altered to confer dominant LoF properties are shown in cyan. The catalytic E642 residue is labelled magenta. ChimeraX (http://www.rbvi.ucsf.edu/chimerax/) was used to generate the structures shown.