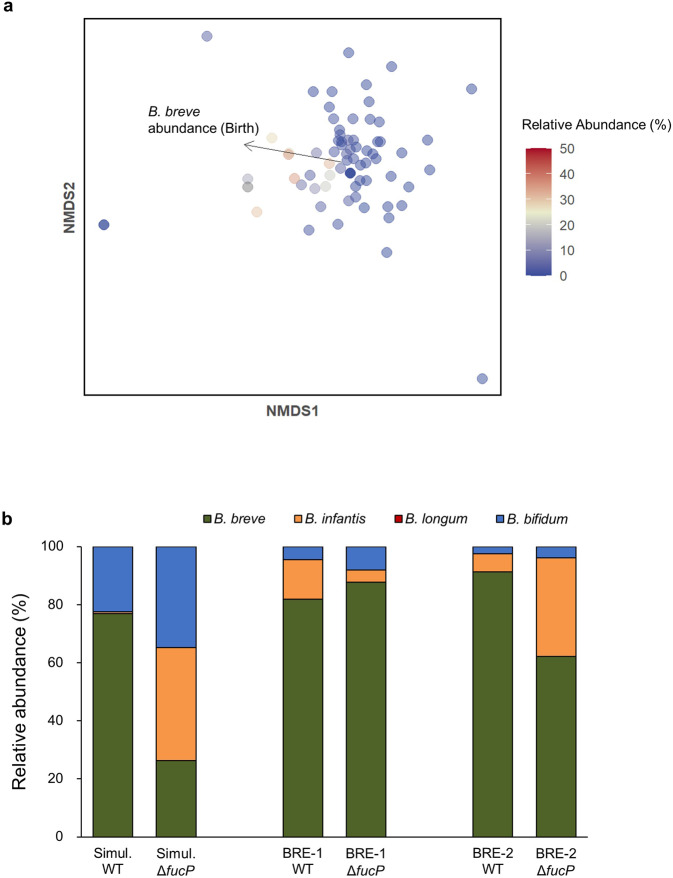
Fig. 4. In silico data mining and in vitro culturing analyses demonstrate that B. breve benefits from priority effects.
a Analysis of in vivo microbiome data. Taxonomic identification was performed using Kraken2 v2, and species abundances were estimated using Bracken v2.6.1. Nonmetric multidimensional scaling (NMDS) ordination plots of bifidobacterial community data from the guts of 4-month old infants who were at least partially breastfed (exclusively breastfed or mixed fed) at birth and at 4 months of age (n = 73) (see Extended Data Fig. 6 and Supplementary Table 5). Each point corresponds to one individual. The color gradient indicates the B. breve abundance within the total gut microbiota at 4 months of age. Statistically significant loadings are indicated as black arrows. b Culturing results using the ΔfucP mutant of B. breve UCC2003. Sequences of 4-species culturing in which B. breve dominated was used, and the experiment was repeated with the wild type (WT) and ΔfucP mutant of B. breve UCC2003. Relative abundance of each species was quantified using qPCR, and each strain is indicated by different colors (blue: B. bifidum, green: B. breve, orange: B. infantis, red: B. longum). Data represent averages of biological triplicates.