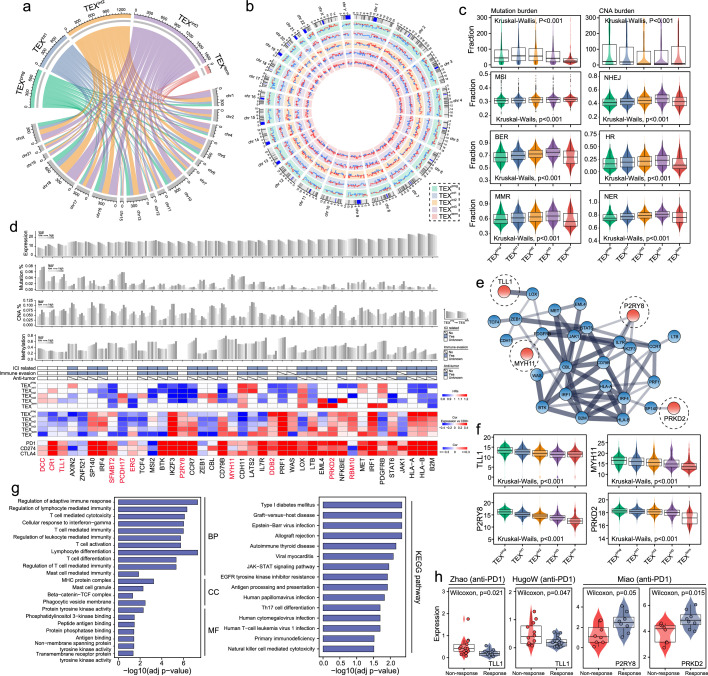
Figure 4.
Featured multi-omic alterations in TEX-based pan-cancer subgroups. (a-b) Circular plots showing the differences in single-nucleotide polymorphism and copy number variations among the five TEX subgroups. (c) Box plots displaying the differences in mutation burden, CNA burden, MSI, BER, MMR, NHEJ, HR and NER among the five TEX subgroups. (d) Gene-level summary of the multi-omics features in the pan-cancer samples. Bar plots show the expression, somatic mutation alterations, DNA copy number alterations and DNA methylation of a list of crucial TEX-associated driver genes. Light gray represents low levels, whereas dark gray represents high levels. Heatmap shows the hazard ratios of key TEX-associated driver genes, the correlation between the expression of these TEX-related driver genes and the fraction of CD8+ T cells, PD-1, PD-L1 and CTLA-4 in the five TEX subgroups. (e) The protein–protein interaction network is formed using the key TEX-associated driver genes. (f) Box plots displaying the expression levels of TLL1, MYH11, P2RY8 and PRKD2 among the five TEX subgroups. (g) Bar charts showing enriched gene ontology terms and KEGG pathways. (h) Box plots showing the expression levels of TLL1, P2RY8 and PRKD2 in responders and non-responders following immunotherapy in the datasets indicated. TEX, T cell exhaustion; CNA, copy number alterations; MSI, microsatellite instable; BER, base excision repair; MMR, mismatch repair; NHEJ, non-homologous end joining; HR, homologous recombination; NER, nucleotide excision repair; PD-1, programmed cell death protein 1; PD-L1, programmed cell death protein ligand 1; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; TLL1, tolloid 1; MYH11, myosin heavy chain 11; P2RY8, P2Y receptor family member 8; PRKD2, protein kinase 2; KEGG, Kyoto Encyclopedia of Genes and Genomes.