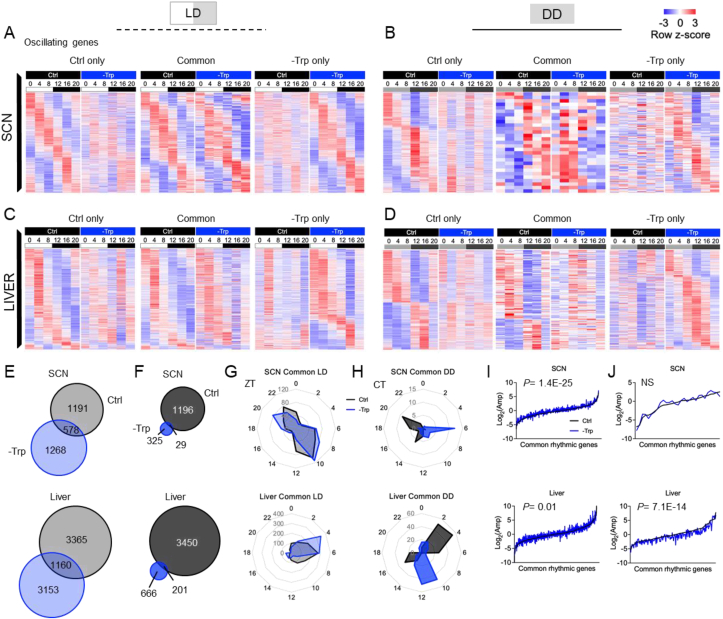
Figure 4.
Light and tryptophan coordinate circadian transcriptional rhythms. A-D. Heatmaps represent the oscillating genes in the SCN (A, B) and the liver (C, D) that are common or unique for the dietary groups of mice housed under light/dark cycles (A, C) or constant darkness (B, D). E-F. Venn diagrams represent the overlap of oscillating genes between the dietary intervention groups in the SCN and the liver in mice housed under light/dark cycles (E) or constant darkness (F). G-H. Radar plots represent the phase of the oscillating metabolites of the SCN and liver in mice housed under light/dark cycles (G) or constant darkness (H). I-J. Amplitude for each gene oscillating in both dietary groups under light/dark conditions (I) or constant darkness (J) plotted in ascending order based on the amplitude in the control groups. Significant oscillations were determined by JTK-CYCLE P < 0.05. The amplitudes were statistically compared between dietary groups by a paired students t-test followed by Bonferroni correction. N = 3 mice. NS = not significant.