Figure 1.
RNA editing and ADAR2 regulate PODXL alternative splicing
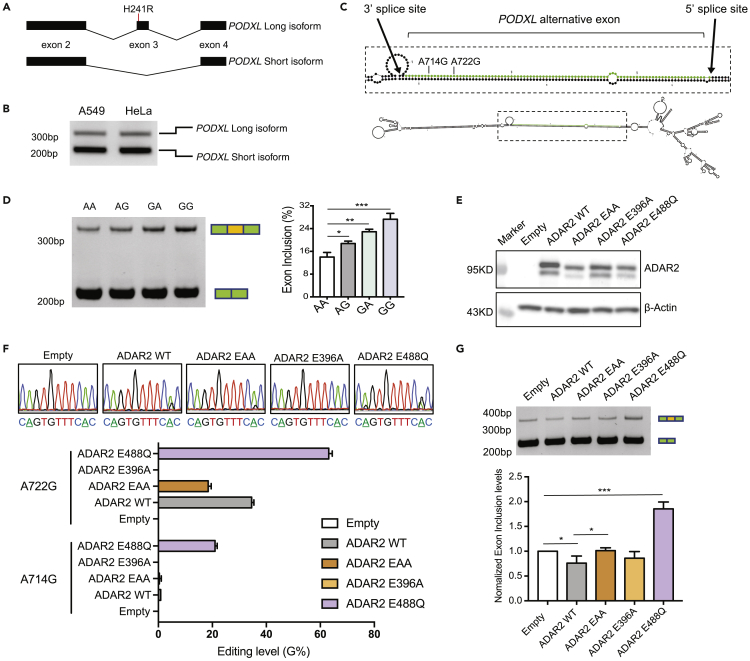
(A) The long and short isoforms of PODXL. Partial gene structures (exon 2 ∼ 4) are shown. The H241R recoding event is labeled in the alternative exon (exon 3) of the long isoform.
(B) Agarose gel image of the endogenous PODXL PCR products amplified from the cDNA of A549 and HeLa cells, respectively.
(C) Predicted RNA structure of the PODXL alternative exon (green) with its flanking introns (black). Locations of the two RNA editing sites are labeled.
(D) Left: PAGE gel resolving the PCR amplicons of transcripts derived from the PODXL splicing reporters in HeLa cells with four combinations of the A714G and A722G editing events (AA, AG, GA, GG, G represents edited and A, unedited). In the AG reporter, G was introduced at the A722G site. In the GA reporter, G was introduced at the A714G site. Right: Quantification of the PODXL alternative exon inclusion rate for each reporter based on the PAGE gel result (measured by ImageJ). Three biological replicates were included. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). See also Figure S1A.
(E) Western blot showing overexpression of ADAR2 and its mutants in HeLa cells. The upper bands represent the FLAG-ADAR2 fusion proteins (see also Figure S1B). The lower bands represent ADAR2 or mutant proteins without FLAG tagging, which may result from alternative translation start sites in the overexpression constructs.
(F) Top: Sanger sequencing to detect RNA editing of the A714G and A722G editing sites (underlined As) in the minigene reporters after co-transfection with ADAR2 overexpression vectors or the empty control in HeLa cells. Bottom: Quantification of the RNA editing levels based on the Sanger sequencing peaks (measured using 4Peaks). Three biological replicates were included. Data are plotted as mean ± SEM. See also Figure S1C.
(G) Top: PAGE gel image of the amplicons of PODXL transcripts derived from the splicing reporters co-transfected with the ADAR2 overexpression vectors or the empty control in HeLa cells. Bottom: Normalized exon inclusion levels based on the PAGE gel band intensity (measured by ImageJ). Three biological replicates were included. For each replicate, the exon inclusion levels were normalized against the empty control. Data are represented as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗∗p < 0.001).