Figure 3.
PODXL isoforms regulate cell migration to different degrees
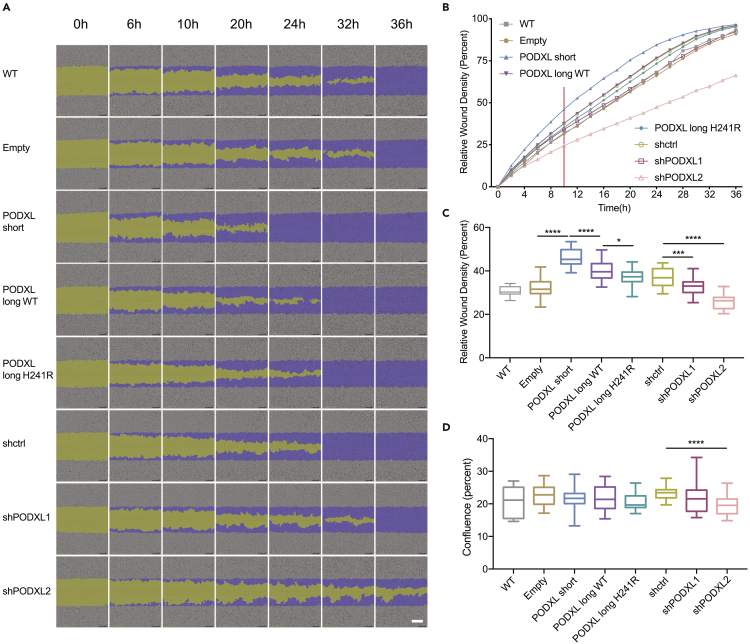
(A) Cell migration assay of A549 cells overexpressing PODXL isoforms or with PODXL knockdown (PODXL-OE/KD A549 cells). WT: wild-type A549 cells. Empty: A549 cells overexpressing the empty backbone. shctrl: A549 cells with scrambled control shRNA. Two alternative shRNAs for PODXL were used (shPODXL1, shPODXL2). Phase-contrast images at different time points were shown (purple: initial scratch wound mask, yellow: wound region). Scale bar: white line at the bottom right, 300 μm. See also Figure S3.
(B) Cell migration curve of the PODXL-OE/KD A549 cells described in (A). The plot shows the average values of one set of experiments performed with three biological replicates. Red vertical line highlights data at 10 h post wound creation.
(C) Quantification of cell migration with relative wound density for the PODXL-OE/KD A549 cells. Data at 10 h post wound creation are shown, which was the earliest time point when we observed significant differences in cell migration among PODXL isoforms. Two independent sets of experiments were performed with three biological replicates included in each experiment. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(D) Quantification of cell proliferation using cell confluence levels for the PODXL OE/KD A549 cells. Data at 10 h post wound creation are shown, to exclude the possible effect of cell proliferation on cell migration differences shown in C. Two independent sets of experiments were performed with three biological replicates included in each experiment. Data are plotted as mean ± SEM. The p-values were calculated using Student’s t-test (∗∗∗∗p < 0.0001). See also Figure S3.