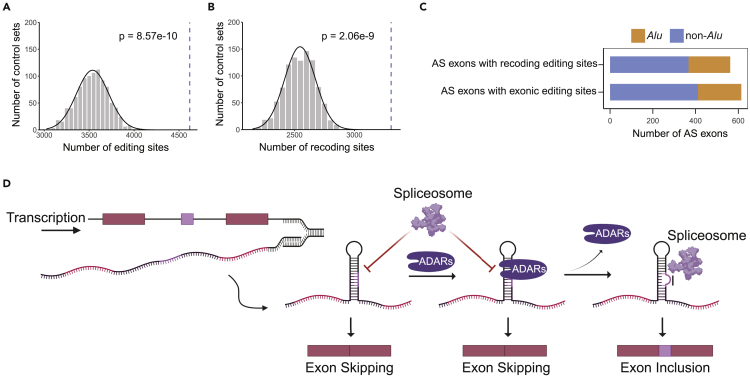
Figure 6.
Exonic editing sites are enriched in alternatively spliced exons
(A) Number of exonic editing sites from REDIportal overlapping alternative exons (blue dashed line), compared to the numbers of exonic editing sites in 1000 sets of random control exons (gray histogram). Black curve represents the normal distribution fit to the histogram. The p-value was calculated using the normal fit.
(B) Similar to A, but for recoding exonic editing sites only.
(C) Number of editing-containing alternative exons overlapping with Alu elements. See also Figure S5.
(D) The multifaceted model of the impact of ADARs/RNA editing on alternative splicing. In the pre-mRNA transcript, the alternative exon forms dsRNA structures with the flanking introns. ADARs bind to the dsRNA structure, which may compete with the spliceosome and prevent splicing. On the other hand, ADARs introduce RNA editing sites to the transcript that destabilize the dsRNA structure. The edited pre-mRNA thus allows access of the spliceosome to the splice sites of the alternative exon, promoting exon inclusion. The illustration was created using BioRender.com.