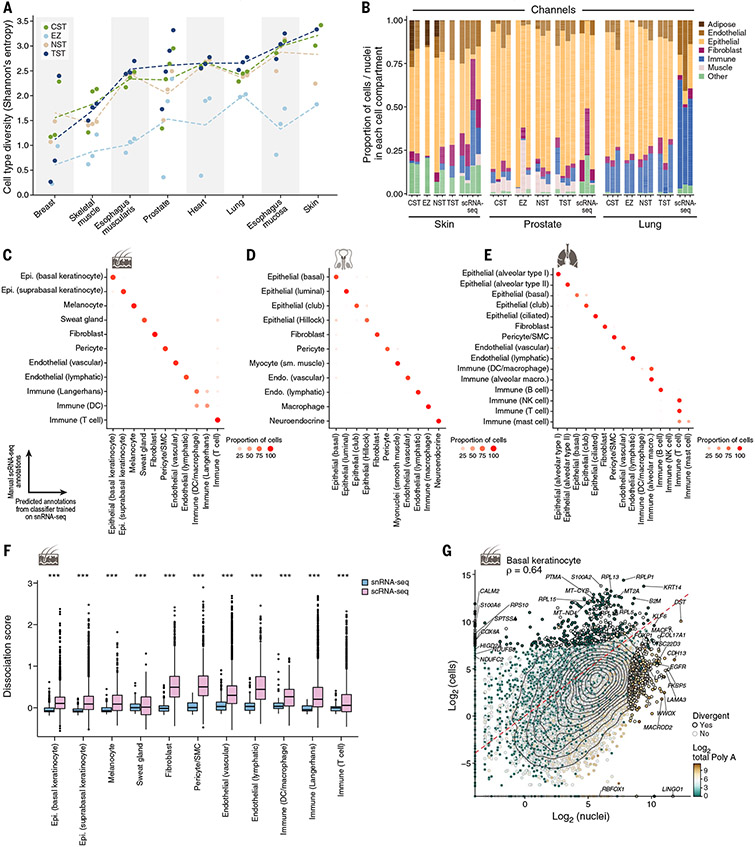
Fig. 2. Concordance of cell-type diversity and cell-intrinsic profiles between snRNA-seq and scRNA-seq.
(A) Cell-type diversity (Shannon entropy, y axis) of each protocol (color) in each sample (dot) and tissue (x axis). Dashed lines indicate the average across samples. (B) Differences in cell proportions. The proportions (y axis) of cells from major categories (color) in each individual by tissue and protocol (x axis) are shown. (C to E) Concordance of cell-intrinsic programs. Proportions of cells (dot color and size) of a manually annotated group (rows) predicted to belong to a given nucleus profile annotation label (columns) by a random forest classifier trained on nuclei and applied to cells of the same tissue for skin (C), lung (D) or prostate (E) are shown. (F) Tissue dissociation expression signatures in scRNA-seq. Scores [y axis, average background corrected log(TP10K+1)] of a dissociation-related stress signature (41) in scRNA-seq (pink) and snRNA-seq (blue) profiles in each major lung cell type (x axis) are shown (***Benjamini-Hochberg FDR < 10−16, Wilcoxon rank sum test). Box plots show median, quartiles, and whiskers at 1.5 times the interquartile range (IQR). (G) Divergent genes between cell and nucleus profiles. Averaged pseudobulk expression (28) of protein-coding genes (dots) in skin basal keratinocyte nuclei (x axis) and cells (y axis) is shown. Divergent genes are represented by a black dot outline. The color scale shows the total length of polyA stretches with at least 20 adenine bases in log2 scale. Epi., epithelial; sm., smooth; SMC, smooth muscle cell.