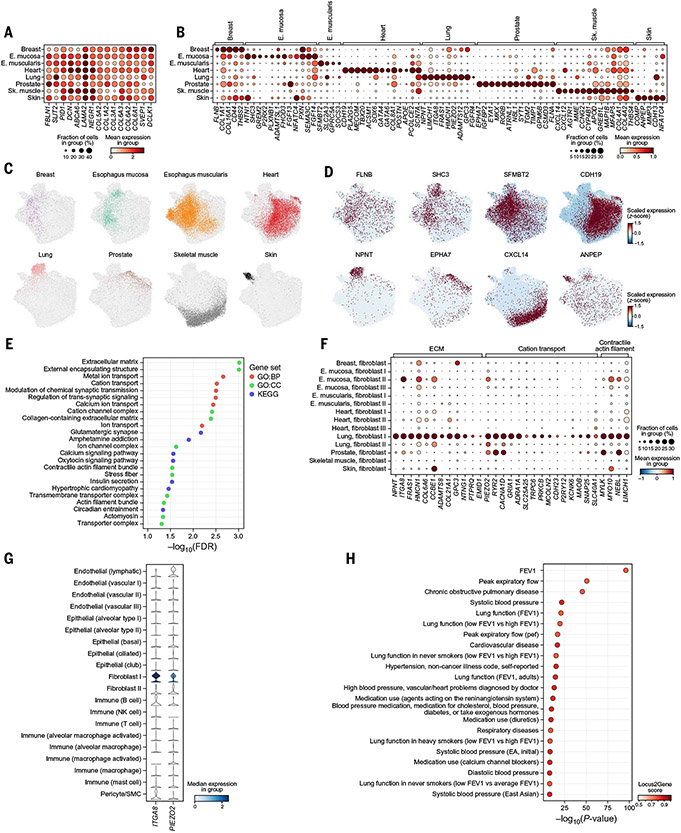
Fig. 4. Shared and tissue-specific fibroblast features.
(A and B) Expression in each tissue subset (rows) of marker genes (columns) distinguishing fibroblasts from nonfibroblasts across all tissues (A) or enriched in fibroblasts in one versus other tissues (B). (C and D) Fibroblast profiles (dots) colored by tissue (C) or expression of the most exclusive marker (D). (E) Significance [−log10(FDR), x axis] of gene sets (y axis) enriched (FDR < 5%) in genes covarying with the lung-specific fibroblast signature. (F) Expression of ECM and cation transport genes (columns) in the covarying gene module in each granular fibroblast subtype in each tissue (rows). (G) ITGA8 and PIEZO2 (columns) expression in granular cell types (rows) in lung. (H) Significance (x axis) and Open Targets Genetics locus-to-gene score (color) of the most significant variants mapped to NPNT with a high (>0.5) locus-to-gene score in GWASs (y axis). FEV, forced expiratory volume.