Abstract
Since the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic began 2 years ago, the scientific community has swiftly worked to understand the transmission, pathogenesis, and immune response of this virus to implement public health policies and ultimately project an end to the pandemic. In this perspective, we present our work identifying SARS-CoV-2 epitopes to quantify T-cell responses and review how T cells may help protect against severe disease. We examine our prior studies which demonstrate durable humoral and cell-mediated memory in natural infection and vaccination. We discuss how SARS-CoV-2–specific T cells from either natural infection or vaccination can recognize emerging variants of concern, suggesting that the currently approved vaccines may be sufficient. We also discuss how pre-existing cross-reactive T cells promote rapid development of immune memory to SARS-CoV-2. We finally posit how identifying SARS-CoV-2 epitopes can help us develop a pan-coronavirus vaccine to prepare for future pandemics.
Keywords: immune memory, epitopes, variants, pre-existing cross-reactive
Identification of SARS-CoV-2 epitopes permit quantification of T cells. Immunity is relatively durable after natural infection and vaccination. SARS-CoV-2–specific T cells can recognize variants of concern. Individuals with pre-existing cross-reactive SARS-CoV-2–specific T cells quickly mount robust immune responses.
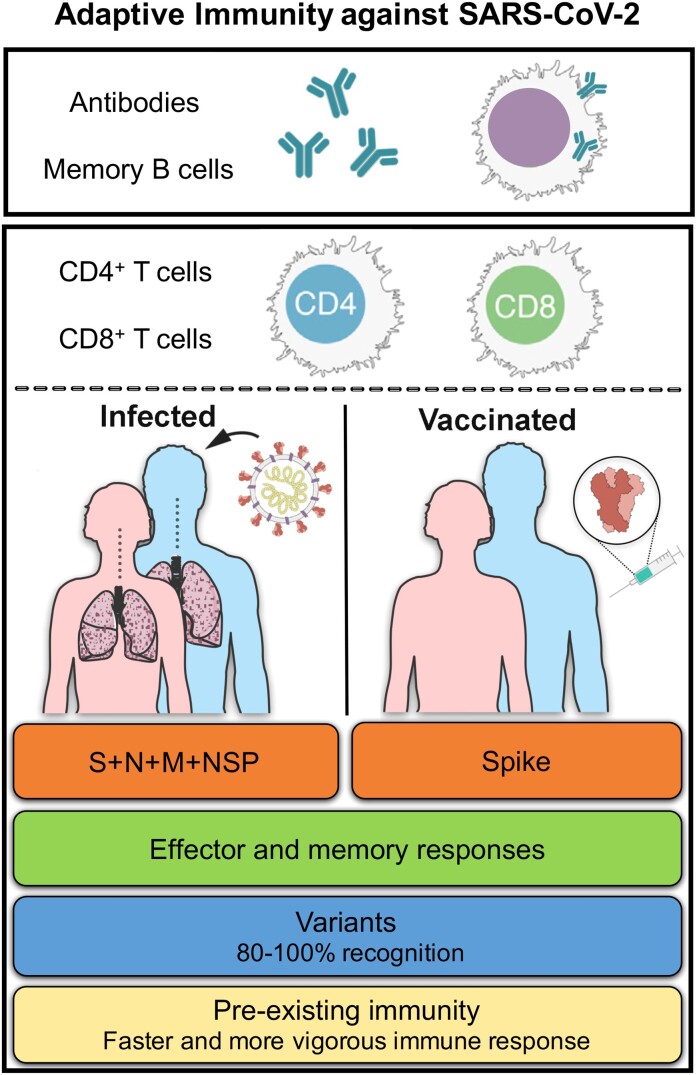
Components of adaptive immunity include antibodies, memory B cells, and CD4+ and CD8+ T cells. In this perspective piece, we present an overview of our studies characterizing the nature and targets of the adaptive responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants, in the setting of natural immunity and vaccination (Figure 1). We further elaborate on the implications of these findings in light of the SARS-CoV-2 pandemic as of October 2021. Finally, we discuss the phenomenon of pre-existing immune memory and its potential implications in the heterogeneity of who develops clinical disease, and boosting of vaccine immune responses.
Figure 1.
Measurable correlates of protection: adaptive immune responses to SARS-CoV-2 infection and vaccination. Abbreviations: M, membrane; N, nucleocapsid; NSP, nonstructural protein; S, spike; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
TARGETS OF ADAPTIVE IMMUNITY IN CONVALESCENT CASES AT THE ANTIGEN LEVEL
In the early stages of the pandemic, we and others performed analyses based on the available SARS-CoV-2 sequences, which highlighted significant homology in the sequence and predicted structure of the antigens encoded in its proteome with other coronaviruses, particularly SARS-CoV and Middle East respiratory syndrome–related coronavirus (MERS) [1]. Previous studies defined B- and T-cell immunogenic regions of the SARS-CoV proteome, which were relatively well conserved in SARS-CoV-2 [2]. Based on these analyses, we predicted and synthesized SARS-CoV-2–specific epitopes, as well as sets of SARS-CoV-2 overlapping peptides spanning the entire proteome, to measure T-cell responses [3].
We first defined adaptive immune responses in self-resolving cases of SARS-CoV-2 infection that did not require hospitalization. This was important as there were no data at that time on adaptive immune responses to SARS-CoV-2. Early in the pandemic, there was a concern that T-cell responses may actually be responsible for severe coronavirus disease 2019 (COVID-19) cases [4]. There was also a concern that adaptive immunity against coronaviruses were weak and thus casted doubt on the prospects for vaccine development [5, 6].
To this end, we measured antibody and CD4+ and CD8+ T-cell responses. Neutralizing antibodies are key components of adaptive responses, induced by the vast majority of vaccines, that bind and neutralize a virus and thus have the capacity to prevent infection. We fully expected antibodies to play an important role. T cells are not usually associated with prevention of infection, but instead have the capacity to recognize and eliminate infected cells that are not accessible by antibodies, thus limiting disease and clearing an infection. Several studies in animal models indicated that CD4+ T cells were potentially important in controlling other coronavirus infections. We also hypothesized that CD8+ T cells were important in limiting and/or resolving infection, as they can recognize and eliminate infected cells.
We detected antibody responses in all subjects and strong CD4+ and CD8+ T-cell responses in the majority of convalescent subjects [3]. Dominant responses were directed against the spike (S) antigen for both CD4+ and CD8+ T cells, as well as antibodies. The fact that S was dominantly recognized by all 3 arms of the adaptive immune response was an exciting result, supporting the ongoing development of S-based vaccines [7–10]. This was not fully expected, based on the pattern of responses in other viral families, where surface antigens are often dominant antigens for antibody responses, while core/nonstructural antigens are often dominant targets for T-cell recognition [11, 12]. We also detected good T-cell responses against the membrane (M), nucleocapsid (N), and nonstructural protein (NSP) antigens, raising the possibility that targeting these antigens may provide a greater breadth of T-cell responses, and that conserved regions in these proteins may target different coronaviruses and SARS-CoV-2 variants.
DEFINING THE EPITOPE REPERTOIRE RECOGNIZED BY CD4+ AND CD8+ T CELLS
Subsequent studies detailed the repertoire of epitope specificities recognized in a cohort of 99 COVID-19 convalescent subjects [13]. These results confirmed previous studies mapping SARS-CoV-2 immunodominant antigens [3]. At the individual level, they also highlighted that the response is multi-specific and prominently recognizes 3–4 different antigens for both CD4+ and CD8+ T-cell responses.
To define CD4+ T-cell epitopes, we used an unbiased approach, testing overlapping 15-mers spanning the entire SARS-CoV-2 genome. In the case of CD8+ T-cell epitopes, we tested epitopes predicted to bind to 28 different HLA-A and B alleles, providing coverage for the majority of alleles expressed in the general population, irrespective of ethnicity. The study identified 280 and 523 different CD4+ and CD8+ T-cell epitopes, respectively [13]. We inferred that each subject was able to recognize 15–20 different epitopes per each T-cell population. Furthermore, because T-cell recognition is HLA restricted, and because of the high degree of polymorphism of HLA molecules, the specific epitopes recognized would typically differ from one individual to the next. This large repertoire of epitopes, recognized by both SARS-CoV-2–specific CD4+ and CD8+ T cells, suggests that it would be unlikely that SARS-CoV-2 could escape T-cell recognition at the population level [14].
The epitope identification studies also underscored HLA polymorphism as an important source of response heterogeneity [13]. Certain HLA class I alleles mediate strong responses, focused on relatively few dominant epitopes, and conversely, other alleles restrict responses mediated by many epitopes, each associated with a more moderate magnitude of response [13]. HLA polymorphism has been shown to influence immunity and disease outcomes in several diseases, and future investigations will establish whether this also applies to SARS-CoV-2 [15].
Over time our group developed and refined different epitope and peptide pools that can be used to accurately and robustly measure immune responses. We have been proactive in sharing these reagents with the research community worldwide. Current efforts are aimed at generating pools of epitopes with immunodiagnostic value that can accurately determine the prior vaccination and/or infection history of a given individual subject. These reagents will allow us to determine which subjects were neither infected nor vaccinated, only vaccinated, only infected, and both infected and vaccinated.
IMMUNE MEMORY FOLLOWING SARS-CoV-2 INFECTION
In the early days of the pandemic, there was uncertainty about protection from reinfection, and the possibility was raised that there might be poor immune memory to the SARS-CoV-2 virus, and/or to coronaviruses in general. Therefore, we decided to directly measure the 4 main components of immunological memory. These are as follows: (1) antibodies; (2) memory B cells that can produce more antibodies after infection; (3) CD4+ T cells that are necessary for the generation of neutralizing antibodies, can help boost subsequent antibody responses, and have direct antiviral activities themselves; and (4) CD8+ T cells, the classic killer T cells that kill virally infected cells.
Immune memory post–SARS-CoV-2 infection was studied in a cohort of almost 200 individuals, the largest cohort for which memory over time was measured for any acute viral infection [16]. We followed immune responses over 8 months. This was necessary because the kinetics of human immune memory were not well defined, and at early time points immune responses can follow a pattern of marked expansion and contraction. Responses at 6 months or longer provide a better view of the kinetics of immune memory after infection or vaccination.
The vast majority of subjects were positive for spike immunoglobulin G (IgG) and neutralizing antibodies up to 8 months postinfection, consistent with other large studies that examined serological endpoints [17–19]. Memory B cells to the spike receptor binding domain (RBD) or nucleocapsid were more abundant at 3 to 6 months postinfection than at 1 month postinfection, which was probably the biggest surprise [16]. This is consistent with additional work by Michel Nussenzweig and others [20, 21]. In the case of CD8+ T cells, approximately 70% and 50% of individuals were positive for memory at early and at later time points, respectively. The half-life observed was comparable to studies analyzing CD8+ T-cell responses to a high-quality yellow fever vaccine [22]. For CD4+ T cells, nearly 100% of individuals tested positive 1 month postinfection, and approximately 90% remained positive at 8 months postinfection [16].
The overall picture emerging when serology, memory B cell, memory CD8+, and memory CD4+ T cells are considered at the same time and in the same people is a complex one, with each compartment following different kinetics. Significant heterogeneity was detected from one individual to the next, with 100-fold differences in the different memory compartments. At 6 months postinfection, approximately 95% of individuals were still positive for different components of immune memory [16]. This overall picture was confirmed by other laboratories, including a large study by Juliana McElrath and Rafi Ahmed [23].
This relatively stable memory suggests that the majority of individuals are likely to be protected at least from severe SARS-CoV-2 reinfections for extended periods of time, possibly for years into the future. This is consistent with observational studies finding that previously infected people are resistant to reinfections for at least 6 to 8 months [24, 25].
CORRELATES OF PROTECTION FROM COVID-19 INFECTION
The simplest option for any vaccine to protect against infection and disease is the development of high-level, long-lasting neutralizing antibodies. It is likely that antibodies, in particular neutralizing antibodies, play the dominant role in vaccine protection from infection, while T cells limit disease severity. In short, these arms of the adaptive immune system are important and work together. Accordingly it is likely that optimal vaccine design stimulates both humoral and cellular immunity.
It is reasonable to posit that hospitalization-level COVID-19 can be prevented by generating a combination of antibodies, memory B cells, CD4+ T cells, and CD8+ T cells. Several lines of evidence point to protective contributions of T cells against COVID-19. We found that a coordinated and early adaptive immune response in the acute phase of disease is linked to more favorable outcomes [26]. Furthermore, we observed no convincing evidence of causal negative associations of adaptive immunity with disease severity [26, 27]. Early T-cell responses correlate with better outcomes and lower viral loads in SARS-CoV-2 infection [28]. Similarly, CD8+ T cells can provide control of SARS-CoV-2 in nonhuman primates, whereas dysregulated CD8+ T-cell responses contribute to lung pathology in severe disease [29, 30]. Moreover, agammaglobulinemic and B-cell–depleted individuals have only a moderately increased risk of hospitalization with COVID-19 [31, 32]. Finally, 1 dose of a Moderna or Pfizer mRNA vaccine provided substantial protection in the absence of detectable neutralizing antibodies in most individuals [33].
The SARS-CoV-2 virus replicates very quickly in nasal spaces and oral cavities, with an incubation period of approximately 2 days [34]. Severe disease and hospitalizations are typically associated with viral shedding in a variety of tissues and for more days than in mildly ill patients [35]. The differential kinetics of this virus in different tissues provide an opportunity for memory T and B cells to contribute to virus control after initial infection. However, additional layers of immunity may contribute to protection, particularly when high levels of neutralizing antibodies might not be present, have waned, or their potency has decreased against variants of concern. In the simplest terms, it is a race between the virus and the immune system. Vaccines change the dynamics of that race, giving the immune system a head start.
IMMUNE MEMORY FOLLOWING mRNA VACCINATION
More recent studies focused on immune memory associated with COVID-19 vaccines. A recent study examined samples from a clinical trial of the Moderna mRNA-1273 vaccine [36]. While both a low dose of 25 µg and a dose of 100 µg were assessed in the initial clinical trial, the 100-µg dose was selected for Emergency Use Authorization. We asked how immune memory to the 25-µg dose compared with the 100-µg dose, which is of general relevance in a dose-sparing perspective. We also wanted to evaluate T-cell memory for an RNA vaccine in general, as there was a big gap in our understanding of immune memory induced by these vaccines, which clearly elicit good antibody response and disease protection [36].
Accordingly, we measured antibodies and CD4+ and CD8+ T cells over a 7-month span, including 6 months after the second immunization. The results were consistent with reported serology for the mRNA-1273 vaccine. The 25-µg dose behaved similarly to the 100-µg dose, with comparable kinetics and quality of memory although moderately lower amounts [36]. For T-cell memory to the 25-µg dose, there was only a 2-fold decline in the magnitude of CD4+ T cells between peak and 6 months. At the 6-month post–second immunization time point, almost 100% of individuals still had measurable CD4+ T-cell memory. Notably, there was significant T-follicular helper (Tfh) memory, a key point that had not been addressed for mRNA-based vaccines before. CD8+ T-cell responses were detected in almost 90% of subjects at early time points, declining to about two-thirds of subjects at the 6-month time point. The decay of the CD8+ T-cell memory was similar to CD4+ T-cell memory, suggesting it would likely be durable for years to come [36]. While the 100-µg dose induced approximately 2-fold higher antibody and CD4+ T-cell responses than the 25-µg dose, CD8+ T-cell responses were equivalent. Based on these results, dose sparing for this vaccine seems plausible. In terms of a comparison between the RNA vaccine and infection, we found that the 25-µg dose produced antibody and CD4+ and CD8+ T-cell responses to spike comparable to natural infection [36].
Age is a major factor in terms of potential waning of immunity. As of 1 April 2022, an additional vaccination is now recommended for individuals age 12 years or older. We found that older subjects, either age 55 years or older or age 70 years or older, had 2-fold reduced peak antibodies and antibodies at 7 months, consistent with findings with the 100-µg mRNA-1273 dose. Our study was the first to address T-cell memory, with T-cell memory being at least as good in older individuals, and actually trending higher [36].
NEGLIGIBLE IMPACT OF VARIANT-ASSOCIATED MUTATIONS ON T-CELL RECOGNITION
A recent meta-analysis inventoried the SARS-CoV-2 T-cell epitopes recognized in humans and new studies are constantly curated in the Immune Epitope Database (IEDB) [2, 37]. As of 10 October 2021, 53 different studies are curated in the IEDB, describing, in total, 1863 different epitopes. As previously noted, the remarkably high number of T-cell epitopes recognized by human responses against SARS-CoV-2 indicates that it would be unlikely for the virus to be able to escape T-cell recognition at the population level.
Further bioinformatical analysis confirmed this notion, and showed that in excess of 90% of all CD4+ and CD8+ T-cell epitopes are totally (100%) conserved in variants of concern, including Alpha, Beta, and Gamma [14]. This was the case irrespective of whether the whole proteome was considered, or if the analysis was limited to the spike antigen, relevant in the context of potential escape of vaccine-induced responses. Parallel experimental analysis with overlapping peptides spanning the entire spike antigen, or the entire proteome, shows that the T-cell recognition of these variants was essentially not impacted, and 80–100% of T-cell recognition was maintained for the different variants tested, for both CD4+ and CD8+ T-cell responses [14]. This was verified in the case of spike responses from vaccinated individuals, or responses to spike and other components of the viral proteome in naturally infected individuals.
We also noted that, while the median reactivity at the population level was not largely impacted, some subjects were associated with a more than 2 times decrease in CD4+ or CD8+ T-cell responses [14]. It is possible that these outliers may be individuals expressing particular HLA types, mediating a dominant response, that is also mediated at least in part by epitopes affected by variant mutations [21].
In summary, the data indicate that T-cell reactivity is largely preserved in the cases of SARS-CoV-2 variants, including Delta. These data were confirmed by 2 recent independent studies [38, 39]. Despite relatively substantial decreases in neutralizing antibody responses to SARS-CoV-2 variants, a largely intact T-cell response may play a role in controlling infection and preventing more severe outcomes.
THE ROLE OF PRE-EXISTING CROSS-REACTIVE IMMUNITY ON IMMUNE RESPONSES TO INFECTION AND VACCINATION
In the spring of 2020, we and several other groups noted that immune reactivity to SARS-CoV-2 sequences could be detected in people that had not been exposed to SARS-CoV-2 [3, 27, 40–43]. Reactivity was detected in samples banked from 2015–2018, excluding prior SARS-CoV-2 exposure. It was hypothesized that this pre-existing immunity may be linked to prior exposure to common cold coronaviruses with immune memory recognizing epitopes that shared significant homology with SARS-CoV-2.
A subsequent paper defined the epitope repertoire of pre-existing memory T cells recognizing both SARS-CoV-2 and homologous common cold coronavirus peptides [40]. In several cases, common cold coronavirus sequences were recognized better than SARS-CoV-2 sequence, implying that the original epitope that elicited the response was likely a common cold coronavirus epitope. Whether pre-existing memory T-cell activity was associated with a functional consequence remained open to debate. While some feared that cross-reactivity might be deleterious, invoking a possible “antigenic sin” phenomenon, we and others hypothesized that pre-existing memory may be beneficial, allowing for a kinetic advantage in the generation of immune responses in natural infection or vaccination [44, 45].
Samples from the 25-µg mRNA-1273 vaccine trial described above provided an opportunity to directly test this hypothesis of T-cell functionality. We classified each individual based on whether they had pre-existing T-cell reactivity at baseline, before their first immunization. Analysis of samples from subsequent time points revealed that individuals with pre-existing immunity were able to mount a faster and more vigorous CD4+ T-cell response against the spike protein. Importantly, pre-existing immune memory was also associated with better antibody responses [36]. It was further shown that the presence of these cross-reactive T cells was linked to favorable outcomes in healthcare worker cohorts followed longitudinally over time, and also possibly linked to abortive infection [46, 47].
Additional studies illustrate that pre-existing immunity is associated with better outcomes in vaccination and natural infection [48–50]. This effect might contribute to the heterogeneity in clinical outcomes in natural infection, and provides indirect but powerful evidence in favor of T-cell reactivity being relevant to the development of protective immunity to SARS-CoV-2. However, there are likely more immunologic factors than pre-existing immunity that could predict whether someone develops severe disease. In particular, the interplay between innate and adaptive immunity is complex and is likely to influence disease outcomes.
THE POTENTIAL FOR ELICITING BROAD-BASED CORONAVIRUS T-CELL RESPONSES AS A VACCINE STRATEGY
The data discussed above are compatible with the notion that T-cell responses might be associated with milder disease outcomes, and that pre-existing cross-reactive immunity can have a positive influence on the outcome of SARS-CoV-2 infection. This re-emphasizes the potential for broadening the repertoire of T-cell responses induced by vaccination by including additional antigens, antigen fragments, or epitopes.
In this context, it is possible to hypothesize that research efforts could identify sequences broadly conserved across different coronavirus species, including coronaviruses of potential concern from zoonotic sources, and known or predicted sequences that are targets for human T-cell responses. Current studies are characterizing immune responses against 2 common cold coronaviruses, OC43 and NL63, and detailing the antigens and epitopes recognized. These data will help define components of immune responses that are conserved across different sarbecoviruses, more generally in Beta coronaviruses, or even conserved across Alpha and Beta coronaviruses. This strategy could be exploited to generate a vaccine that is broadly reactive, not only against the SARS-CoV-2 and its variants but potentially other coronaviruses that could jump into humans in the future and generate a new pandemic.
Contributor Information
Jennifer Dan, La Jolla Institute for Immunology, La Jolla, California, USA; Department of Medicine, Division of Infectious Diseases and Global Public Health, La Jolla, California, USA.
Ricardo da Silva Antunes, La Jolla Institute for Immunology, La Jolla, California, USA.
Alba Grifoni, La Jolla Institute for Immunology, La Jolla, California, USA.
Daniela Weiskopf, La Jolla Institute for Immunology, La Jolla, California, USA.
Shane Crotty, La Jolla Institute for Immunology, La Jolla, California, USA; Department of Medicine, Division of Infectious Diseases and Global Public Health, La Jolla, California, USA.
Alessandro Sette, La Jolla Institute for Immunology, La Jolla, California, USA; Department of Medicine, Division of Infectious Diseases and Global Public Health, La Jolla, California, USA.
Notes
Acknowledgments. These studies are the result of a close collaboration between several laboratories and core facilities at the La Jolla Institute and laboratories at several other institutions, including the groups of Davey Smith at University of California, San Diego (UCSD), Richard Scheuerman at J. Craig Venter Institute (JCVI), Simon Mallal at Vanderbilt University, Florian Krammer at Mount Sinai, and many others. A particular heartfelt thank you to La Jolla Institute for Immunology’s Clinical, Cytometry and Bioinformatics core and to all donors who selflessly donated blood for our studies.
Financial support. This work was supported by the National Institutes of Health (grant numbers K08 A135078 to J. D., 75N9301900065 to D. W. and A. S., U01CA260541-01 to D.W., and AI142742 to A. S. and S. C.).
Supplement sponsorship. This supplement is sponsored by the Precision Vaccines Program of Boston Children's Hospital.
References
- 1. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 2020; 27:671–80.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vita R, Mahajan S, Overton JA, et al. . The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 2019; 47:D339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grifoni A, Weiskopf D, Ramirez SI, et al. . Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020; 181:1489–501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peeples L. News feature: avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc Natl Acad Sci U S A 2020; 117:8218–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choe PG, Perera RAPM, Park WB, et al. . MERS-CoV antibody responses 1 year after symptom onset, South Korea, 2015. Emerg Infect Dis 2017; 23:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Okba NMA, Raj VS, Widjaja I, et al. . Sensitive and specific detection of low-level antibody responses in mild middle east respiratory syndrome coronavirus infections. Emerg Infect Dis 2019; 25:1868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Polack FP, Thomas SJ, Kitchin N, et al. . Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020; 383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baden LR, Sahly HME, Essink B, et al. . Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2020; 384:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sadoff J, Gray G, Vandebosch A, et al. . Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med 2021; 384:2187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keech C, Albert G, Cho I, et al. . Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N Engl J Med 2020; 383:2320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weiskopf D, Angelo MA, de Azeredo EL, et al. . Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci U S A 2013; 110:E2046–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greenbaum JA, Kotturi MF, Kim Y, et al. . Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci U S A 2009; 106:20365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tarke A, Sidney J, Kidd CK, et al. . Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med 2021; 2:100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tarke A, Sidney J, Methot N, et al. . Impact of SARS-CoV-2 variants on the total CD4 + and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2021; 2:100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol 2018; 18:325–39. [DOI] [PubMed] [Google Scholar]
- 16. Dan JM, Mateus J, Kato Y, et al. . Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021;371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wajnberg A, Amanat F, Firpo A, et al. . Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 2020; 370:1227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Egbert ER, Xiao S, Colantuoni E, et al. . Durability of spike immunoglobin G antibodies to SARS-CoV-2 among health care workers with prior infection. JAMA Netw Open 2021; 4:e2123256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gudbjartsson DF, Norddahl GL, Melsted P, et al. . Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 2020; 383:1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaebler C, Wang Z, Lorenzi JCC, et al. . Evolution of antibody immunity to SARS-CoV-2. Nature 2021; 591:639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Z, Muecksch F, Schaefer-Babajew D, et al. . Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature 2021; 595:426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akondy RS, Johnson PLF, Nakaya HI, et al. . Initial viral load determines the magnitude of the human CD8 T cell response to yellow fever vaccination. Proc Natl Acad Sci U S A 2015; 112:3050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cohen KW, Linderman SL, Moodie Z, et al. . Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med 2021; 2:100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lumley SF, O’Donnell D, Stoesser NE, et al. . Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med 2020; 384:533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crotty S. Hybrid immunity. Science 2021; 372:1392–3. [Google Scholar]
- 26. Moderbacher CR, Ramirez SI, Dan JM, et al. . Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020; 183:996–1012, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weiskopf D, Schmitz KS, Raadsen MP, et al. . Phenotype and kinetics of SARS-CoV-2–specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 2020; 5:eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tan AT, Linster M, Tan CW, et al. . Early induction of functional SARS-CoV-2 specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Reports 2021; 34:108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McMahan K, Yu J, Mercado NB, et al. . Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021; 590:630–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheon IS, Li C, Son YM, et al. . Immune signatures underlying post-acute COVID-19 lung sequelae. Sci Immunol 2021; 6:eabk1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soresina A, Moratto D, Chiarini M, et al. . Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immu 2020; 31:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Apostolidis SA, Kakara M, Painter MM, et al. . Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat Med 2021; 27:1990–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kalimuddin S, Tham CYL, Qui M, et al. . Early T cell and binding antibody responses are associated with Covid-19 RNA vaccine efficacy onset. Med (N Y) 2021; 2:682–8.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lauer SA, Grantz KH, Bi Q, et al. . The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 2020; 172:577–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Y, Zhang L, Sang L, et al. . Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest 2020; 130:5235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mateus J, Dan JM, Zhang Z, et al. . Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science 2021; 374:eabj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grifoni A, Sidney J, Vita R, et al. . SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe 2021; 29:1076–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alter G, Yu J, Liu J, et al. . Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature 2021; 596:268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Geers D, Shamier MC, Bogers S, et al. . SARS-CoV-2 variants of concern partially escape humoral but not T cell responses in COVID-19 convalescent donors and vaccine recipients. Sci Immunol 2021; 6:eabj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mateus J, Grifoni A, Tarke A, et al. . Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020; 370:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meckiff BJ, Ramírez-Suástegui C, Fajardo V, et al. . Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell 2020; 183:1340–53.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bert NL, Tan AT, Kunasegaran K, et al. . SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020; 584:457–62. [DOI] [PubMed] [Google Scholar]
- 43. Braun J, Loyal L, Frentsch M, et al. . SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020; 587:270–4. [DOI] [PubMed] [Google Scholar]
- 44. Sette A, Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol 2020; 20:457–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lipsitch M, Grad YH, Sette A, Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol 2020; 20:709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Swadling L, Diniz MO, Schmidt NM, et al. . Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022; 601:110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kundu R, Narean JS, Wang L, et al. . Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat Commun 2022; 13:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. da Silva Antunes R, Pallikkuth S, Williams E, et al. . Differential T cell reactivity to endemic coronaviruses and SARS-CoV-2 in community and health care workers. J Infect Dis 2021; 224:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sagar M, Reifler K, Rossi M, et al. . Recent endemic coronavirus infection is associated with less severe COVID-19. J Clin Invest 2020; 131:e143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loyal L, Braun J, Henze L, et al. . Cross-reactive CD4 + T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science 2021; 374:eabh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]