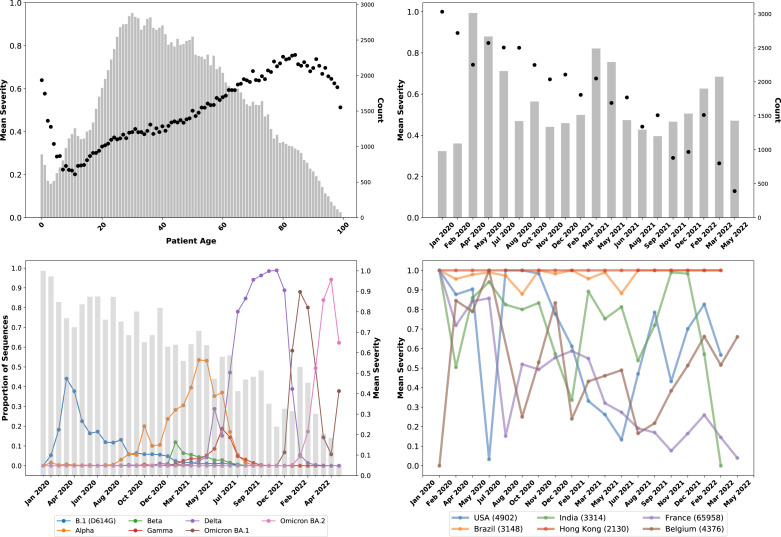
Fig. 1.
Overview of age, sample collection date, and country metadata trends in GISAID data.(A – Upper Left) Mean case severity, where 0 is Mild and 1 is Severe, which equates to the probability of a severe case) by patient age in the GISAID database. The bars show the count of samples for each age. Increasing age trends with increasing severity, as expected, with differences at extremely low and old ages characterized by low sample counts. (B – Upper Right) Mean clinical severity (probability of severe case) by sample collection date recorded in the GISAID data. For clarity, data have been binned over time periods; the bars indicate the number of samples. Over time, the proportion of severe cases has declined, although that trend has been less consistent since Fall 2021. (C – Lower Left) Proportion of sequences in the GISAID patient data set (sequences with patient metadata) for principal variants, including B.1 (the ancestral lineage with the D614G which emerged in Northern Italy and New York in February–March 2020) and its sublineages, Alpha (B.1.1.7 and “Q” sublineages), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2 and AY sublineages), and the two major Omicron lineages, BA.1 and BA.2 (and their sublineages). The bars indicate the mean case severity for each date bin. The trends of sequential lineage waves in GISAID patient data appear to be consistent with the larger GISAID data set, i.e., showing successive Alpha, Delta, and Omicron waves. (D – Lower Right) Mean case severity of samples separated by GISAID metadata for the country where the sequence was collected for selected countries. The total number of sequences in the GISAID patient data set per country is shown within parentheses in the legend. Fluctuations in severity observed in countries appear due to systemic issues or differences in where samples are collected (e.g., in hospitals or outside settings) at different times.