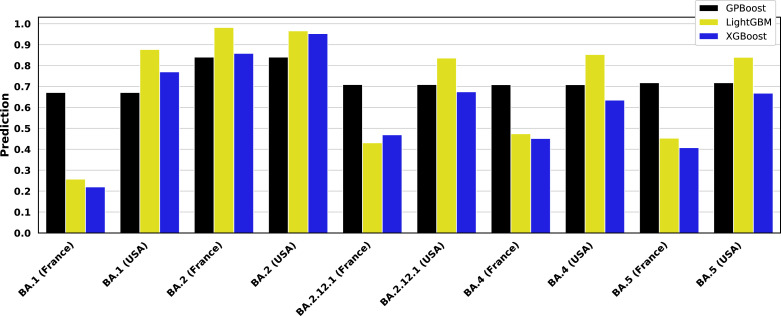
Fig. 7.
Predictions of Omicron subvariant severity. Trained GPBoost, LightGBM, and XGBoost models are simulated for representative BA.1, BA.2, BA.2.12.1, BA.4, and BA.5 sequences from a 60 year-old male patient obtained in the United States, France, and Mexico. The GISAID accession numbers of the sequences for the sequences are: EPI_ISL_6590782 (BA.1), EPI_ISL_7852877 (BA.2), EPI_ISL_12048110 (BA.2.12.1), EPI_ISL_11674447 (BA.4), and EPI_ISL_12029894 (BA.5). The predictions shown here are for models trained on training data as shown in Fig. 4, Fig. 6 where country is a feature for LightGBM and XGBoost. The GPBoost predictions shown here are for the mean of the model response, and it does not vary by country, since country is not a fixed effect in the mixed effects model trained using GPBoost. By contrast, LightGBM and XGBoost predictions fluctuate significantly by simulated country. Emerging Omicron subvariants are uniformly predicted to be more severe than BA.1.