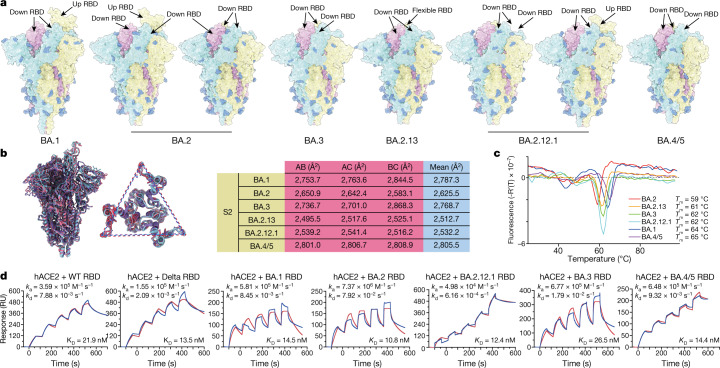
Fig. 1. Structural and receptor-binding characteristics of Omicron subvariants.
a, Surface representation of S-trimers of BA.1, BA.2, BA.3, BA.2.13, BA.2.12.1 and BA.4/BA.5 (BA.4/5) variants. b, Structural interpretation and functional verification of the stability of the spike protein of BA.1, BA.2, BA.3, BA.2.13, BA.2.12.1 and BA.4/BA.5 variants. Left, superimposed structures of spike protein and the S2 domains of BA.1 (purple), BA.2 (red) and BA.4/BA.5 (blue). The binding surface areas between S2 subunits of the variants are calculated in the table on the right. c, Thermoflour analysis for these Omicron variants. Analyses were performed as biological duplicates. d, Binding affinities of RBDs of Omicron variants for hACE2 measured by SPR. Analyses were performed as biological duplicates.