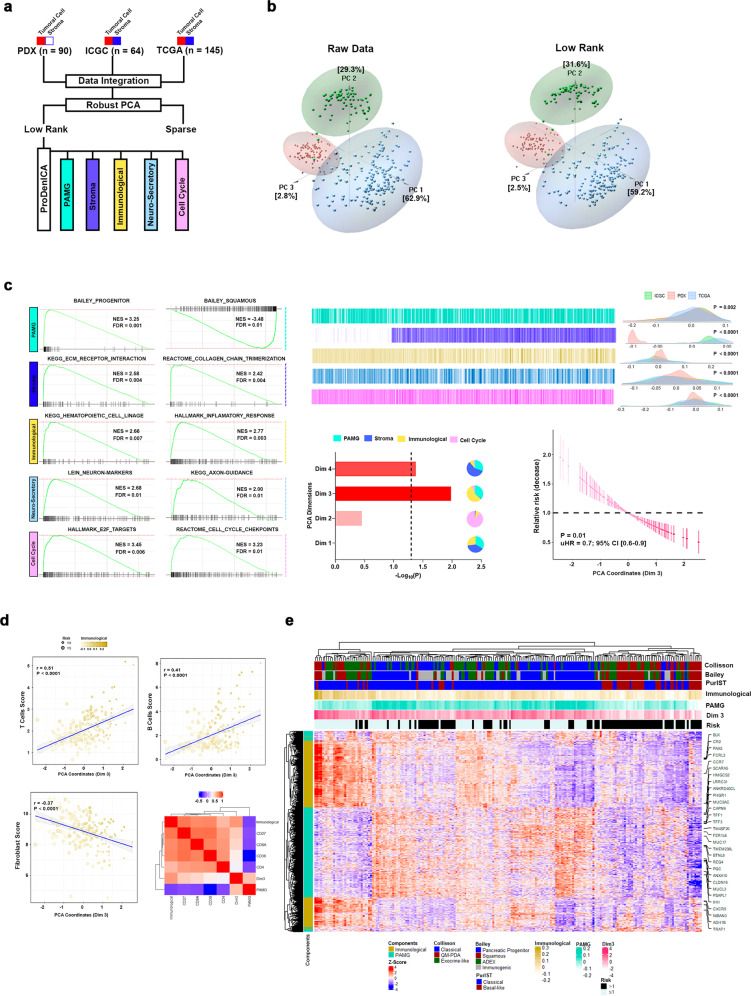
Fig. 1. PDAC biological relevant components determination.
a Low-Rank ICA (LRICA) methodology was applied on the integrated PDX, TCGA-PAAD, and ICGC-PACA-AU Seq datasets to establish the tumor phenotype gradients. b Comparing the original integrated expression matrix with the Low-Rank matrix applying principal component analysis (PCA). c Five PDAC biological relevant components were detected after LRICA, applying as the selection criteria, the kurtosis, and the biological enrichment through GSEA. Also, PCA was applied to evaluate the component contribution to the patient’s prognosis. Dimension 3 (Dim 3) showed the highest association with the prognosis (uHR = 0.76, 95% CI [0.62–0.94]; P = 0.01). This dimension was explained mainly by the PAMG (35.8%) and Immunological (57.2%) components. d T (r = 0.51; P < 0.0001) and B (r = 0.41; P < 0.0001) cells scores after stroma deconvolution of bulk RNA displayed a positive correlation with the Dim 3, whereas the fibroblast (r = −0.37; P < 0.0001) score was negative. This observation was complemented by correlation of the T and B cell markers. Specifically, CD8A and CD27 displayed the highest coefficients. e Heatmap of the high contributive gene of PAMG and Immunological components for TCGA-PAAD, and ICGC-PACA-AU Seq datasets.