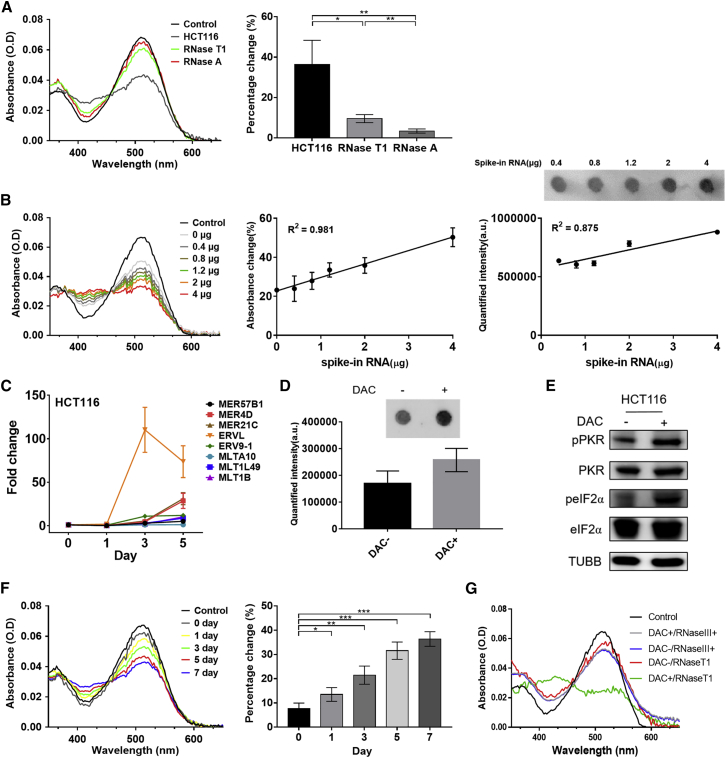
Figure 2.
Detection of total dsRNA from HCT116 cells
(A) The absorbance spectra of Am-MC when mock-treated (HCT116), RNase T1-treated (RNase T1), or RNase A-treated (RNase A) HCT116 total RNA was added. Quantified percentage change at 515-nm peak is shown on the right. (B) Recovery of the indicated amount of EGFP dsRNAs spiked in to 4 μg of total RNA from HCT116 cells. The absorbance spectra of Am-MC for one set of experiment is shown on the left. The middle shows quantified absorbance change for three biological replicates with linear fitting; 400 ng of RNase T1-treated samples were used for the analysis. The right shows the dot blot using a J2 antibody for one set of experiments and the quantified intensity of the biological triplicates; 1 μg of RNase T1-treated RNA was used for the analysis. (C) Selected ERV RNA expression in total RNAs extracted from HCT116 cells after treating them with DAC for the indicated number of days. (D) Dot-blot assay to detect elevated dsRNA expression after DAC treatment; 1 μg of total RNA from HCT116 cells 5 days after the DMSO (DAC−) or DAC treatment was used for the analysis. (E) Western blotting of the PKR signaling pathway on HCT116 cell lysates 5 days after the DMSO (DAC−) or DAC treatment. TUBB was used as a loading control. (F) The absorbance spectra (left) and quantified 515-nm peak (right) of 2 μg of RNase T1-treated total RNAs from HCT116 cells after DAC treatment for the indicated duration. (G) The absorbance spectra of Am-MC when RNAs from DAC-treated HCT116 cells (5 days after the treatment) were digested with RNase III; 2 μg of RNase III digested RNA was used. In all plots, an average of three biological replicates are shown with error bars indicating SD. A Student’s t test was used for pairwise comparison, and one-way ANOVA was also used to analyze the data in (F). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.