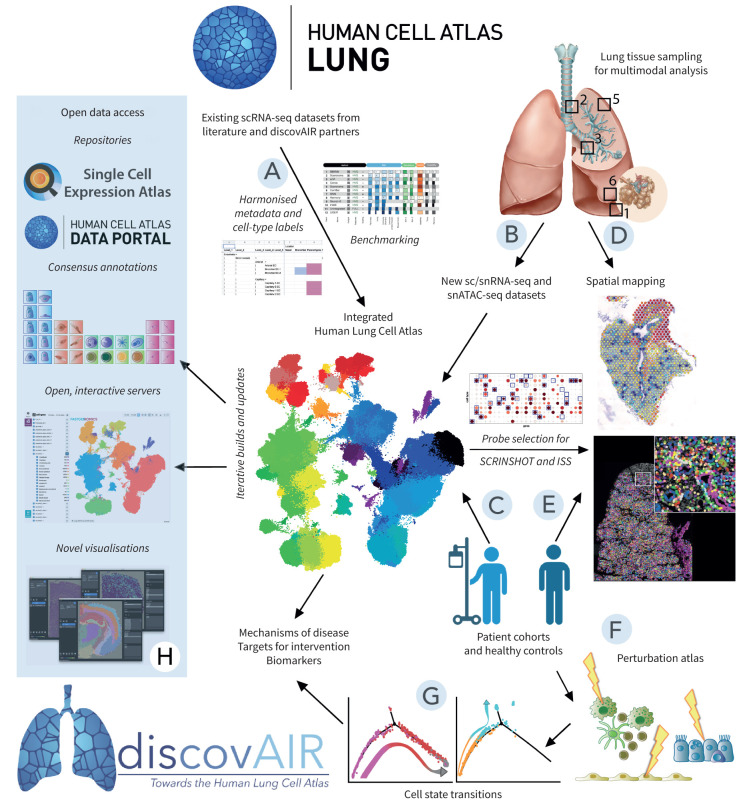
FIGURE 1.
discovAIR approach and workflow. The discovAIR consortium aims to establish a first draft of the Human Lung Cell Atlas by integrating existing single-cell RNA sequencing (scRNA-seq) datasets from the Lung Biological Network of the HCA (HCA-Lung) into a single embedding, allowing analyses on the integrated dataset (A). This dataset will be enriched by additional datasets generated by the discovAIR consortium: a multi-omics characterisation of lung tissue from five healthy donor lungs sampled at five locations (“deep dive”) (B) and a cohort of healthy controls (at least n=50) and patients with lung disease (at least n=50) (C). The “deep dive” tissue samples will also be used to generate spatially resolving datasets on matched tissue sections using Visium, in situ sequencing, SCRINSHOT (single-cell resolution in situ hybridisation on tissues) and laser capture microdissection (LCM)-guided single-nucleus RNA sequencing (snRNA-seq) analysis (D). Samples from the discovAIR cohort (patients and controls) will be used for spatial mapping using SCRINSHOT (E) and for establishment of the lung cell perturbation atlas (F). In the lung cell perturbation atlas, stimulations of ex vivo cultured primary cells or precision-cut lung tissue slices will be used to map cell state trajectories of healthy cells to diseased cell states (G). These novel datasets will be ingested into the next iterations of the integrated Human Lung Cell Atlas, used to establish consensus annotations for the different lung cell types and submitted to the appropriate data repositories (H) as open data using novel visualisation modalities for the spatial datasets. Finally, the integration of the Human Lung Cell Atlas (including datasets from lung disease) and the lung cell perturbation atlas will generate novel insights into mechanisms of disease, and help guide identification of drug targets and biomarkers for disease inception or treatment response. ISS: in situ sequencing; snATAC-seq: single-nucleus sequencing assay for transposase-accessible chromatin.