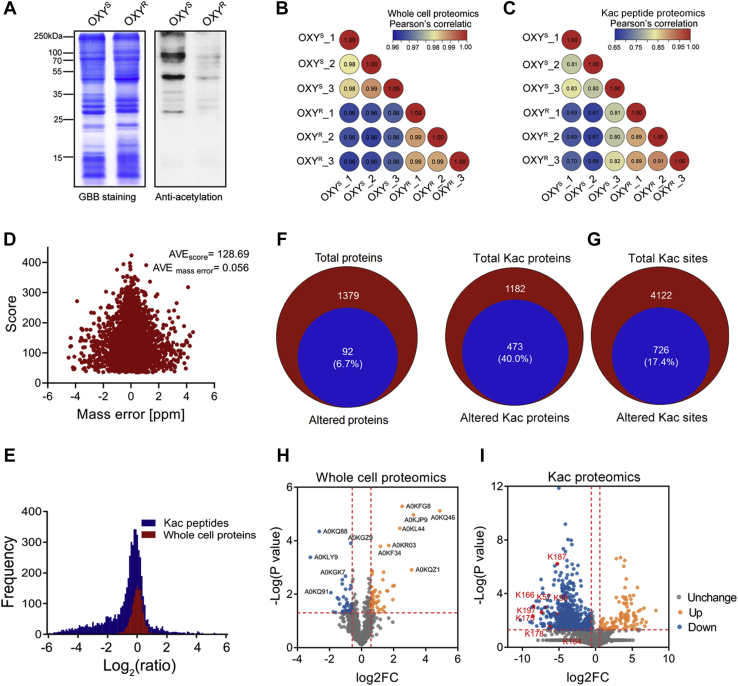
Fig. 1.
Whole-cell and Kac peptide proteomics analysis between OXYSand OXYRstrains.A, the Kac profiles in OXYS and OXYR strains by Western blotting. The left figure is the Coomassie blue staining as loading control. B and C, correlation coefficient analysis of whole-protein and Kac peptide proteomics in three biological repeats for each group, respectively. D, the distribution of mass error and the identified score of Kac-modified peptides. E, the frequency distribution of protein/peptide ratio (OXYRversus OXYS) in both whole-cell protein and Kac-modified peptide proteomics. F, the overlapping proteins between altered and total identified proteins in whole-cell proteomics. G, the overlapping Kac proteins/peptides between altered and total identified Kac proteins/peptides in Kac proteomics. H and I, volcano plot of the p-values versus the log2 protein abundance differences between OXYS and OXYR, with proteins or peptides outside the significance lines colored in orange or blue, respectively. Some representative protein names and Kac sites are highlighted. OXYS, oxytetracycline sensitive; OXYR, oxytetracycline resistance; Kac, lysine acetylation.