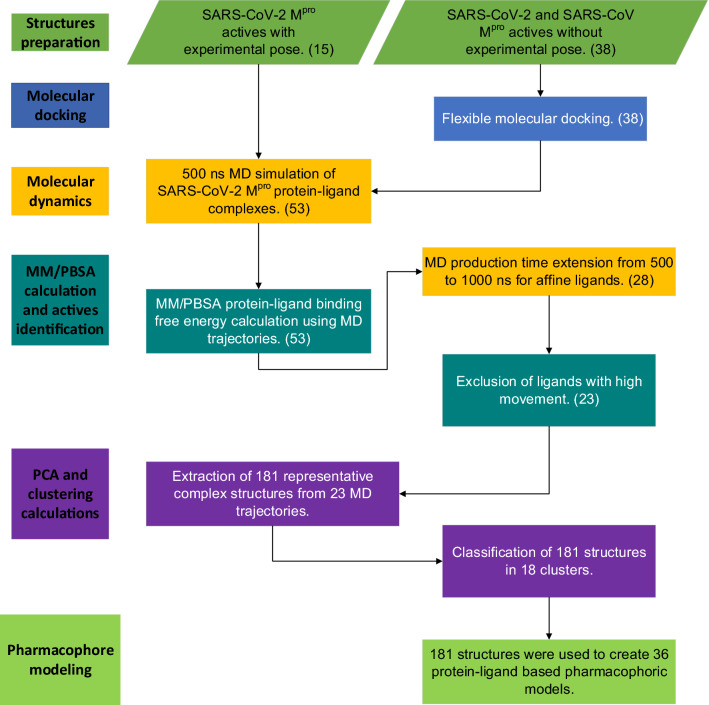
Figure 2.
Summary of the methods to generate pharmacophore models for SARS-CoV-2 Mpro. We collected 15 inhibitors with protein–ligand crystal available and 38 inhibitors with protein–ligand crystal unavailable. These 38 inhibitors went through a molecular docking process to predict ligand poses on the active site. The 38 predicted poses and the 15 experimental poses were taken to perform a MD simulation with 500 ns production time. The MD trajectories were used for MM/PBSA free energy calculations and 28 systems with protein–ligand binding free energy less than − 100 kJ/mol were identified and the respective ligands were considered as possible actives. The MD production time of these 28 systems was extended to 1000 ns. Ligands showing prominent movement on the active site were excluded resulting in 23 actives. The 23 MD trajectories were used for a PCA and clustering process in order to extract representative conformations of each ligand on the active site. Finally, the conformations were employed to create protein–ligand-based pharmacophore models.