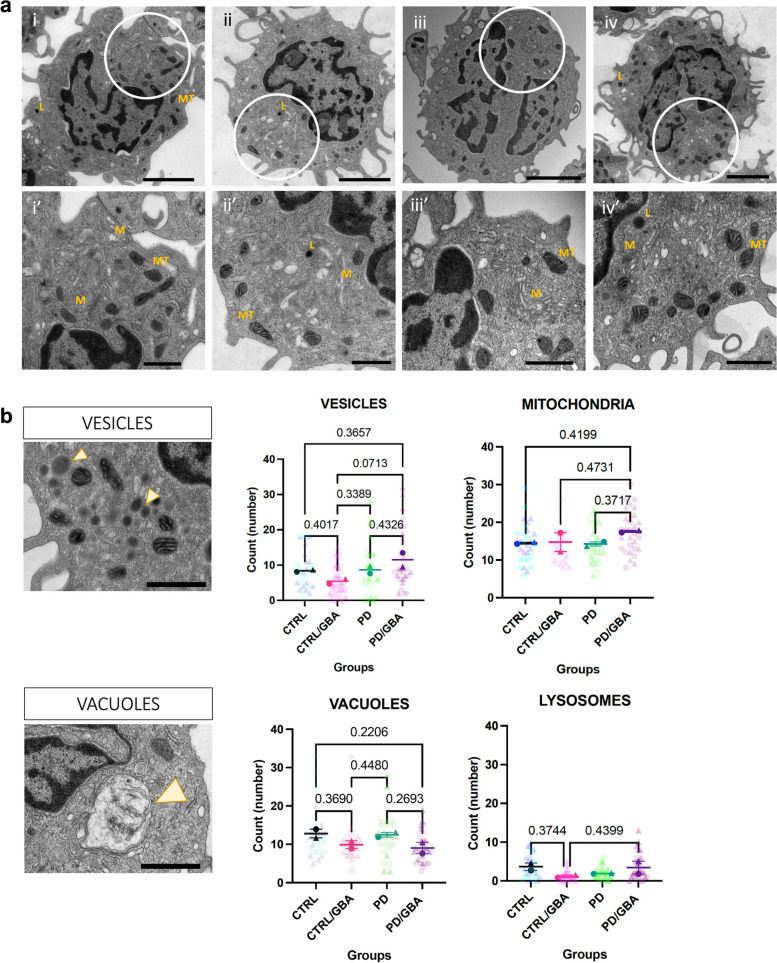
Fig. 5.
Morphological characterization of CD14+ monocytes. a Low magnification (1800X) images (i, ii, iii, iv) and high magnification (4000x) images (i’, ii’, iii’, iv’). Location of the high magnification images within low magnification pictures are highlighted by the white circle in the first row. Scale bar: 2 𝝻m and 800 nm respectively. i/i’) Cells from CTRL and ii/ii’) CTR/GBA subjects showing normal membrane compartment (M, Golgi and Endoplasmic reticulum), lysosomes (L), mitochondria (MT), cell-to-cell adherences, multiple pseudopodal extensions from the cell membrane. iii/iii’) Cells from PD subjects showing highly thickened, and distorted cell membrane compartment (M, Golgi and Endoplasmic reticulum); decreased pseudopodia, RER and free ribosomes; mitochondrial (MT) membranes are severely affected, often lacking external membranous encapsulation and with internally swollen cristae. iv/iv’) Cells from PD/GBA subjects showing small and large vacuoles, normal cell membranes, pseudopodal extensions appear normal, and nuclei. Abnormal membrane assembly (M), ultrastructure, and free ribosomes. b Quantification of mitochondria, lysosome, vesicles and vacuoles in four groups (n = 2 subjects per group, 15 cells per sample). Example of vacuole and vesicles reported in the figures on the left highlighted by yellow arrowhead (scale bar 800 nm). Data represents count from each cell from the two samples per group, and mean and SEM of the two replicates. p-value from one-way ANOVA analysis is reported above each comparison (p-value < 0.5 were reported)