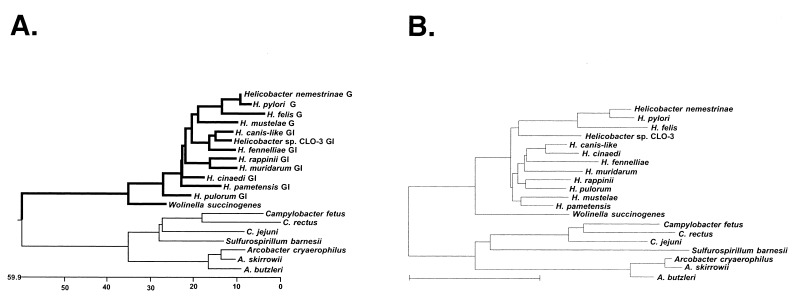
FIG. 2.
(A) Phylogenetic tree of derived amino acid sequences from around the rpoB-rpoC genes junction in ɛ-proteobacteria. The following helicobacters were used in the analysis: H. pylori 26695 (7), H. felis (8), H. nemestrinae (NADC 6904), and H. mustelae (ATCC 43772T) (all gastric; labeled “G” in the figure); H. cinaedi (CCUG 18818T), H. fennelliae (NADC 3235), H. muridarum (NADC 6895), H. canis-like (NADC 29176), H. pametensis (NADC 6900), Helicobacter sp. CLO3 (CCUG 14564), H. rappini (NADC 9615), and H. pulorum (all gastrointestinal; labeled “GI” in the figure). Other organisms used were W. succinogenes (ATCC 29543), C. rectus (NADC 3165), A. skirrowii (NADC 3700), A. cryaerophilus (NADC 3544), A. butzleri (3492), and S. barnesii (SES-3). The deduced amino acid sequences were aligned and the tree was built by using the Clustal method with the PAM250 residue weight table (Windows 32 MegAlign program, version 3.1.7 of DNASTAR). (B) Phylogenetic tree derived from 16S rRNA sequences.