Fig. 2. EV-derived miR-1290 and miR-1246 are highly expressed in metastatic breast cancer patient sera, associated with poor clinical outcomes, enriched in BCBM, and upregulated by tGLI1.
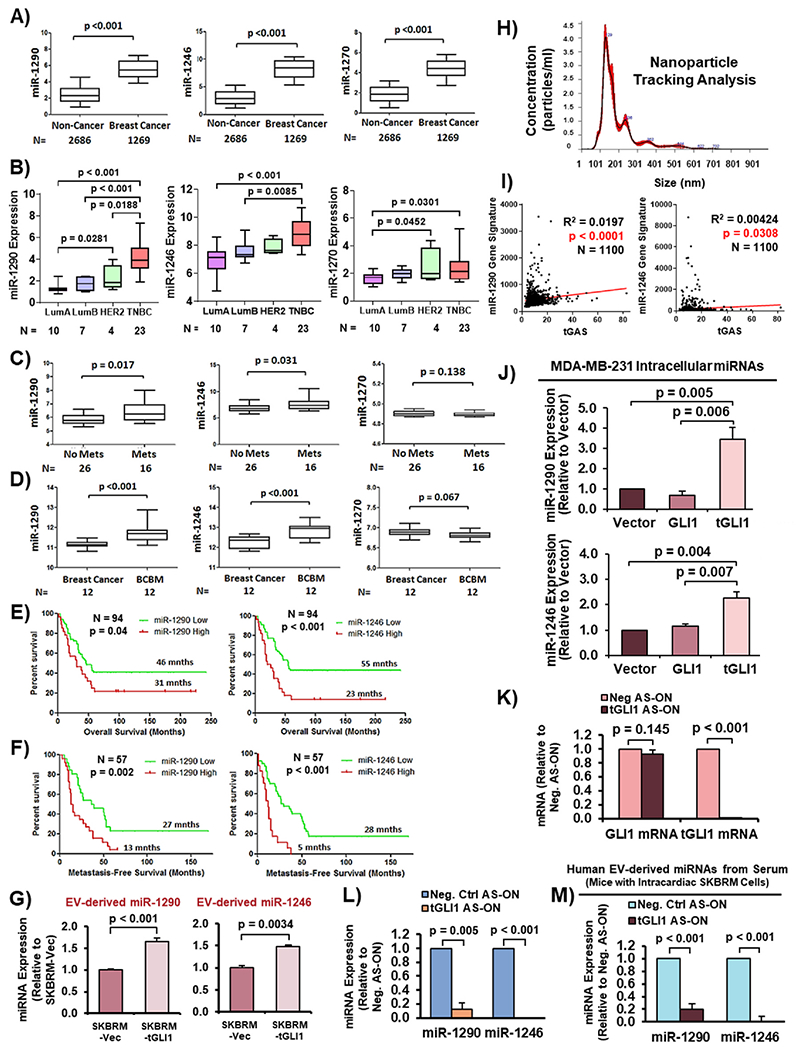
(A) MiRNA expression in sera of healthy individuals (non-cancer) compared to breast cancer patients (GSE73002). (B) Expression of miRNAs in breast cancer patient tumors with known subtype information (GSE86278). LumA, luminal A. LumB, luminal B. TNBC, triple-negative breast cancer. (C) MiRNA expression in sera of breast cancer patients without metastases (No Mets) compared to those with metastases (Mets; GSE68373). (D) MiRNA expression in primary breast tumors and matched brain metastases (GSE37407). (E–F) Kaplan-Meier curves showing overall survival and metastasis-free survival (MFS) of breast cancer patients with low versus high miRNA expression (GSE40267). Log-Rank (Mantle-Cox) test was used to compute p-values. (G) MiRNA expression in EVs from SKBRM-Vector and SKBRM-tGLI1 cells. (H) NTA of isolated EVs. (I) Correlation of tGAS and miR-1290/miR-1246 gene signatures. Pearson correlation was used to calculate p-values. (J) Intracellular expression of miR-1290 and miR-1246 in isogenic MDA-MB-231 cell lines. (K) RT-qPCR to validate effective and selective tGLI1 knockdown by tGLI1 AS-ON in SKBRM cells. (L) tGLI1 knockdown reduced miR-1290 and miR-1246 expression. (M) Expression of EV-derived miR-1290 and miR-1246 in sera of mice carrying SKBRM BCBM with or without tGLI1 knockdown. Serum was harvested from mice 25 days post inoculation. Students t-test was used in Panels A–D, G, and J-M. N = 3 experimental replicates.
