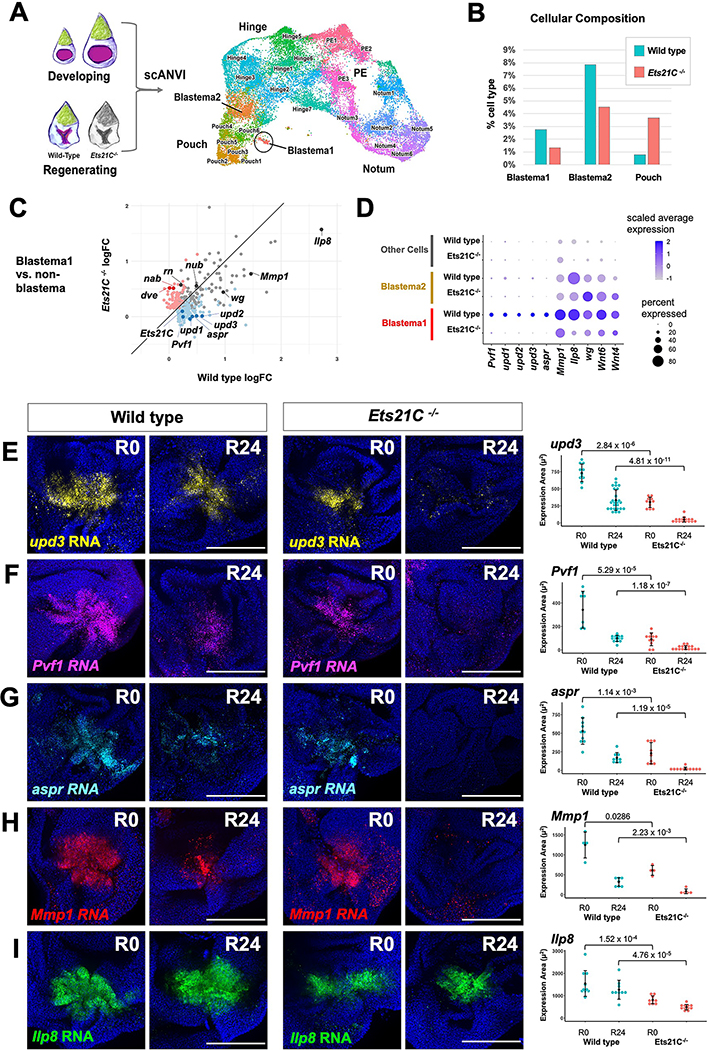
Figure 4. Single-cell analysis reveals genes regulated by Ets21C during regeneration.
(A) Integration of data from regenerating (R24) Ets21C−/− mutant discs with the annotated wild type data shown in Figure 1B. UMAP of harmonized epithelial cells with cell annotations assigned by scANVI. (B) Percent cells assigned to Blastema and Pouch clusters. (C) Natural-log fold-change (logFC) of differentially expressed genes in Blastema1 cells vs non-Blastema cells for wild type versus Ets21C−/− conditions. Blue dots show genes that are significantly (Bonferroni p-value < 0.05) upregulated in Blastema1 cells in wild type but not Ets21C−/−. Red dots highlight genes that are significantly upregulated in Blastema1 cells in Ets21C−/− but not wild type. (D) Dot plot summarizing gene expression for blastema cluster marker genes. (E-I) Wild type and Ets21C−/− mutant discs at early (R0) and mid regeneration (R24) time points with transcripts visualized by HCR as indicated. Area of fluorescence quantified on right with statistical significance between conditions calculated via Wilcoxon test. Microscopy scale bars = 100 μm. See also Figure S4.