Figure 4.
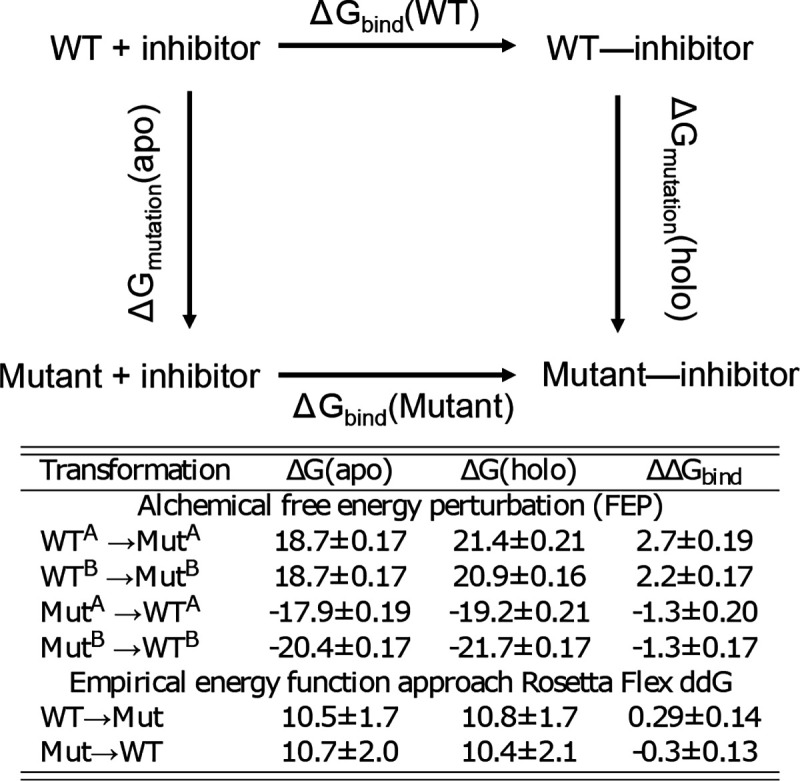
Top. Thermodynamic cycle used to calculate the difference in the noncovalent binding free energy between the mutant and WT Mpros: ΔGbind(Mutant) - ΔGbind(WT)= ΔGmutation(holo) - ΔGmutation(apo). Bottom. FEP and empirical calculations of the change in nirmatrelvir binding free energy of Mpro upon the H172Y mutation. For FEP calculations, transformations were performed on each protomer separately, whereas Rosetta calculations transformed H172 in both protomers simultaneously. ΔΔGbind calculated from the transformation from mutant to WT is less accurate, as it was initiated from the modeled mutant structure. Each calculation was repeated a number of times (see Table S2), the mean and standard errors are reported.
