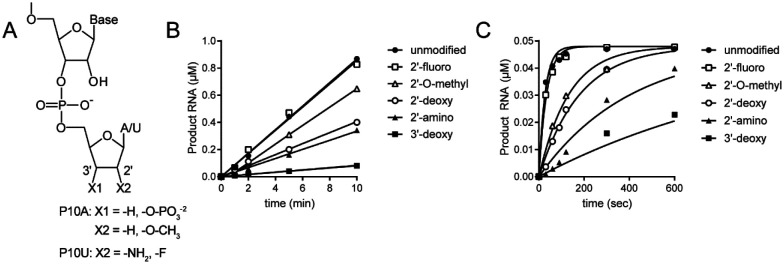
Figure 8. Structure-activity relationships for the ribose of the 3’-terminal nucleotide.
(A) Modifications to the ribose of the terminal nucleotide studied here in the context of P10A: 3’-deoxy (-H), 3’-phosphate (-O-PO3), 2’-deoxy (-H), and 2’-O-methyl (-O-CH3); or P10U: 2’-amino (-NH2) and 2’-fluoro (-F). (B,C) Kinetics of cleavage of modified dsRNAs. In panel B, substrate was present in excess of enzyme. In panel C, enzyme was present in excess of substrate. The data were fit to a line (panel B) or to a single exponential (panel C). Reactions contained 0.1 μM ExoN (1:20) and either 1 or 0.05 μM dsRNA substrate and were quenched at the indicated times. The order in which the modified RNAs were cleaved is as follows: unmodified=2’-F > 2’-O-Me > 2’-deoxy > 2’-amino > 3’-deoxy > 3’-phosphate. The kinetics of cleavage using the 3’phosphate RNA is not shown, because cleavage was not detected. The observed rate of cleavage of modified RNAs and fold difference from the unmodified RNA are shown in Table 3.