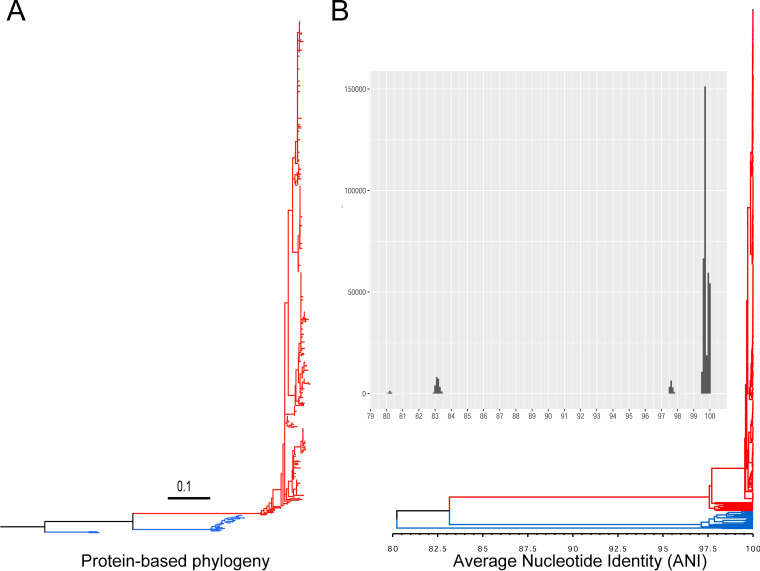
FIG 1.
Taxonomic classification of 634 Brucella genomes downloaded from the NCBI Reference Sequence (RefSeq) database (March 2021). (A) A midpoint-rooted maximum-likelihood (ML) phylogeny of Brucella genomes calculated with PhyloPhlAn. The phylogeny is based on the concatenated alignment of 24,110 amino acid positions of up to 400 universally conserved bacterial proteins. (B) The results of pairwise average nucleotide identity (ANI) between all Brucella genomes calculated with FastANI and plotted with bactaxR. The distribution of the ANI values is represented in the histogram, and the relatedness between all genomes is illustrated by a dendrogram created using the average linkage hierarchical clustering method where the tree height corresponds to ANI similarity. The red branch denotes the genomes of all Brucella species while the blue branches denote divergent genomes from the species B. intermedia (O. intermedium) and Brucella pituitosa, as explained in the text.